Mean deformation metrics for quantifying 3D cell-matrix interactions without requiring information about matrix material properties
- PMID: 26929377
- PMCID: PMC4801239
- DOI: 10.1073/pnas.1510935113
Mean deformation metrics for quantifying 3D cell-matrix interactions without requiring information about matrix material properties
Abstract
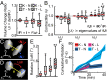
Mechanobiology relates cellular processes to mechanical signals, such as determining the effect of variations in matrix stiffness with cell tractions. Cell traction recorded via traction force microscopy (TFM) commonly takes place on materials such as polyacrylamide- and polyethylene glycol-based gels. Such experiments remain limited in physiological relevance because cells natively migrate within complex tissue microenvironments that are spatially heterogeneous and hierarchical. Yet, TFM requires determination of the matrix constitutive law (stress-strain relationship), which is not always readily available. In addition, the currently achievable displacement resolution limits the accuracy of TFM for relatively small cells. To overcome these limitations, and increase the physiological relevance of in vitro experimental design, we present a new approach and a set of associated biomechanical signatures that are based purely on measurements of the matrix's displacements without requiring any knowledge of its constitutive laws. We show that our mean deformation metrics (MDM) approach can provide significant biophysical information without the need to explicitly determine cell tractions. In the process of demonstrating the use of our MDM approach, we succeeded in expanding the capability of our displacement measurement technique such that it can now measure the 3D deformations around relatively small cells (∼10 micrometers), such as neutrophils. Furthermore, we also report previously unseen deformation patterns generated by motile neutrophils in 3D collagen gels.
Keywords: confocal microscopy; large deformations; neutrophil; traction force microscopy.
Conflict of interest statement
The authors declare no conflict of interest.
Figures







Similar articles
-
3D full-field quantification of cell-induced large deformations in fibrillar biomaterials by combining non-rigid image registration with label-free second harmonic generation.Biomaterials. 2017 Aug;136:86-97. doi: 10.1016/j.biomaterials.2017.05.015. Epub 2017 May 10. Biomaterials. 2017. PMID: 28521203
-
Finite element analysis of traction force microscopy: influence of cell mechanics, adhesion, and morphology.J Biomech Eng. 2013 Jul 1;135(7):71009. doi: 10.1115/1.4024467. J Biomech Eng. 2013. PMID: 23720059 Free PMC article.
-
High resolution, large deformation 3D traction force microscopy.PLoS One. 2014 Apr 16;9(4):e90976. doi: 10.1371/journal.pone.0090976. eCollection 2014. PLoS One. 2014. PMID: 24740435 Free PMC article.
-
Toward single cell traction microscopy within 3D collagen matrices.Exp Cell Res. 2013 Oct 1;319(16):2396-408. doi: 10.1016/j.yexcr.2013.06.009. Epub 2013 Jun 25. Exp Cell Res. 2013. PMID: 23806281 Free PMC article. Review.
-
3D Traction Force Microscopy in Biological Gels: From Single Cells to Multicellular Spheroids.Annu Rev Biomed Eng. 2024 Jul;26(1):93-118. doi: 10.1146/annurev-bioeng-103122-031130. Epub 2024 Jun 20. Annu Rev Biomed Eng. 2024. PMID: 38316064 Review.
Cited by
-
A novel method for sensor-based quantification of single/multicellular force dynamics and stiffening in 3D matrices.Sci Adv. 2021 Apr 9;7(15):eabf2629. doi: 10.1126/sciadv.abf2629. Print 2021 Apr. Sci Adv. 2021. PMID: 33837084 Free PMC article.
-
On the Three-Dimensional Correlation Between Myofibroblast Shape and Contraction.J Biomech Eng. 2021 Sep 1;143(9):094503. doi: 10.1115/1.4050915. J Biomech Eng. 2021. PMID: 33876206 Free PMC article.
-
Flagellar kinematics reveals the role of environment in shaping sperm motility.J R Soc Interface. 2020 Sep;17(170):20200525. doi: 10.1098/rsif.2020.0525. Epub 2020 Sep 9. J R Soc Interface. 2020. PMID: 32900303 Free PMC article.
-
Measurement of dynamic cell-induced 3D displacement fields in vitro for traction force optical coherence microscopy.Biomed Opt Express. 2017 Jan 27;8(2):1152-1171. doi: 10.1364/BOE.8.001152. eCollection 2017 Feb 1. Biomed Opt Express. 2017. PMID: 28271010 Free PMC article.
-
Mechanosensitive traction force generation is regulated by the neutrophil activation state.Sci Rep. 2023 Jul 9;13(1):11098. doi: 10.1038/s41598-023-37997-y. Sci Rep. 2023. PMID: 37423937 Free PMC article.
References
-
- Das T, et al. A molecular mechanotransduction pathway regulates collective migration of epithelial cells. Nat Cell Biol. 2015;17(3):276–287. - PubMed
-
- Discher DE, Janmey P, Wang YL. Tissue cells feel and respond to the stiffness of their substrate. Science. 2005;310(5751):1139–1143. - PubMed
-
- Chen CS. Mechanotransduction - a field pulling together? J Cell Sci. 2008;121(Pt 20):3285–3292. - PubMed
-
- Levental I, Georges PC, Janmey PA. Soft biological materials and their impact on cell function. Soft Matter. 2007;3:299–306. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

