Quantitative genome-wide methylation analysis of high-grade non-muscle invasive bladder cancer
- PMID: 26929985
- PMCID: PMC4854556
- DOI: 10.1080/15592294.2016.1154246
Quantitative genome-wide methylation analysis of high-grade non-muscle invasive bladder cancer
Abstract
High-grade non-muscle invasive bladder cancer (HG-NMIBC) is a clinically unpredictable disease with greater risks of recurrence and progression relative to their low-intermediate-grade counterparts. The molecular events, including those affecting the epigenome, that characterize this disease entity in the context of tumor development, recurrence, and progression, are incompletely understood. We therefore interrogated genome-wide DNA methylation using HumanMethylation450 BeadChip arrays in 21 primary HG-NMIBC tumors relative to normal bladder controls. Using strict inclusion-exclusion criteria we identified 1,057 hypermethylated CpGs within gene promoter-associated CpG islands, representing 256 genes. We validated the array data by bisulphite pyrosequencing and examined 25 array-identified candidate genes in an independent cohort of 30 HG-NMIBC and 18 low-intermediate-grade NMIBC. These analyses revealed significantly higher methylation frequencies in high-grade tumors relative to low-intermediate-grade tumors for the ATP5G2, IRX1 and VAX2 genes (P<0.05), and similarly significant increases in mean levels of methylation in high-grade tumors for the ATP5G2, VAX2, INSRR, PRDM14, VSX1, TFAP2b, PRRX1, and HIST1H4F genes (P<0.05). Although inappropriate promoter methylation was not invariantly associated with reduced transcript expression, a significant association was apparent for the ARHGEF4, PON3, STAT5a, and VAX2 gene transcripts (P<0.05). Herein, we present the first genome-wide DNA methylation analysis in a unique HG-NMIBC cohort, showing extensive and discrete methylation changes relative to normal bladder and low-intermediate-grade tumors. The genes we identified hold significant potential as targets for novel therapeutic intervention either alone, or in combination, with more conventional therapeutic options in the treatment of this clinically unpredictable disease.
Keywords: Epigenetics; HumanMethylation450 BeadChip Array; gene expression; high-grade non-muscle invasive bladder cancer; methylation.
Figures
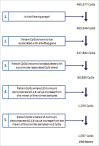
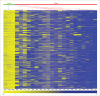
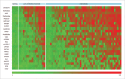


References
-
- Ferlay J, Soerjomataram I, Dikshit R, Eser S, Mathers C, Rebelo M, Parkin DM, Forman D, Bray F. Cancer incidence and mortality worldwide: sources, methods and major patterns in GLOBOCAN 2012. Int J Cancer J Int Du Cancer 2015; 136:E359-86; PMID:26712904; http://dx.doi.org/16399414 10.1002/ijc.29210 - DOI - PubMed
-
- Kirkali Z, Chan T, Manoharan M, Algaba F, Busch C, Cheng L, Kiemeney L, Kriegmair M, Montironi R, Murphy WM, et al.. Bladder cancer: epidemiology, staging and grading, and diagnosis. Urology 2005; 66:4-34; PMID:16399414; http://dx.doi.org/ 10.1016/j.urology.2005.07.062 - DOI - PubMed
-
- Sylvester RJ, van der Meijden AP, Oosterlinck W, Witjes JA, Bouffioux C, Denis L, Newling DW, Kurth K. Predicting recurrence and progression in individual patients with stage Ta T1 bladder cancer using EORTC risk tables: a combined analysis of 2596 patients from seven EORTC trials. Eur Urol 2006; 49:466-5; discussion 75–7; PMID:16442208; http://dx.doi.org/ 10.1016/j.eururo.2005.12.031 - DOI - PubMed
-
- Boustead GB, Fowler S, Swamy R, Kocklebergh R, Hounsome L. Section of Oncology B. Stage, grade and pathological characteristics of bladder cancer in the UK: British Association of Urological Surgeons (BAUS) urological tumour registry. BJU Int 2014; 113:924-30; PMID:24447825; http://dx.doi.org/ 10.1111/bju.12468 - DOI - PubMed
-
- Cambier S, Sylvester RJ, Collette L, Gontero P, Brausi MA, van Andel G, Kirkels WJ, Silva FC, Oosterlinck W, Prescott S, et al.. EORTC Nomograms and Risk Groups for Predicting Recurrence, Progression, and Disease-specific and Overall Survival in Non-Muscle-invasive Stage Ta-T1 Urothelial Bladder Cancer Patients Treated with 1–3 Years of Maintenance Bacillus Calmette-Guerin. Eur Urol 2016; 69(1):60-9; PMID:26210894; http://dx.doi.org/24905984 10.1016/j.eururo.2015.06.045 - DOI - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases
Miscellaneous
