D-BRAIN: Anatomically Accurate Simulated Diffusion MRI Brain Data
- PMID: 26930054
- PMCID: PMC4773122
- DOI: 10.1371/journal.pone.0149778
D-BRAIN: Anatomically Accurate Simulated Diffusion MRI Brain Data
Abstract
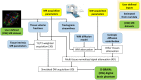
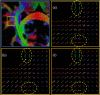
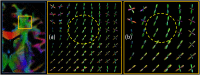
Diffusion Weighted (DW) MRI allows for the non-invasive study of water diffusion inside living tissues. As such, it is useful for the investigation of human brain white matter (WM) connectivity in vivo through fiber tractography (FT) algorithms. Many DW-MRI tailored restoration techniques and FT algorithms have been developed. However, it is not clear how accurately these methods reproduce the WM bundle characteristics in real-world conditions, such as in the presence of noise, partial volume effect, and a limited spatial and angular resolution. The difficulty lies in the lack of a realistic brain phantom on the one hand, and a sufficiently accurate way of modeling the acquisition-related degradation on the other. This paper proposes a software phantom that approximates a human brain to a high degree of realism and that can incorporate complex brain-like structural features. We refer to it as a Diffusion BRAIN (D-BRAIN) phantom. Also, we propose an accurate model of a (DW) MRI acquisition protocol to allow for validation of methods in realistic conditions with data imperfections. The phantom model simulates anatomical and diffusion properties for multiple brain tissue components, and can serve as a ground-truth to evaluate FT algorithms, among others. The simulation of the acquisition process allows one to include noise, partial volume effects, and limited spatial and angular resolution in the images. In this way, the effect of image artifacts on, for instance, fiber tractography can be investigated with great detail. The proposed framework enables reliable and quantitative evaluation of DW-MR image processing and FT algorithms at the level of large-scale WM structures. The effect of noise levels and other data characteristics on cortico-cortical connectivity and tractography-based grey matter parcellation can be investigated as well.
Conflict of interest statement
Figures







References
-
- Le Bihan D, Breton E. Imagerie de diffusion in-vivo par resonance. Cr Acad Sci. 1985;301:1109–1112.
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

