Gtsf1l and Gtsf2 Are Specifically Expressed in Gonocytes and Spermatids but Are Not Essential for Spermatogenesis
- PMID: 26930067
- PMCID: PMC4773171
- DOI: 10.1371/journal.pone.0150390
Gtsf1l and Gtsf2 Are Specifically Expressed in Gonocytes and Spermatids but Are Not Essential for Spermatogenesis
Abstract
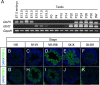
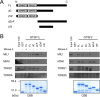
The unknown protein family 0224 (UPF0224) includes three members that are expressed in germ-line cells in mice: Gtsf1, Gtsf1l, and BC048502 (Gtsf2). These genes produce proteins with two repeats of the CHHC Zn-finger domain, a predicted RNA-binding motif, in the N terminus. We previously reported that Gtsf1 is essential for spermatogenesis and retrotransposon suppression. In this study, we investigated the expression patterns and functions of Gtsf1l and Gtsf2. Interestingly, Gtsf1l and Gtsf2 were found to be sequentially but not simultaneously expressed in gonocytes and spermatids. Pull-down experiments showed that both GTSF1L and GTSF2 can interact with PIWI-protein complexes. Nevertheless, knocking out Gtsf1, Gtsf2, or both did not cause defects in spermatogenesis or retrotransposon suppression in mice.
Conflict of interest statement
Figures


References
-
- Abraham LK, Laura LT. Histology and cell biology: An introduction to pathology et al. Elsevier Sauders; 2012. pp. 587–615.
-
- Aravin AA, Sachidanandam R, Girard A, Fejes-Toth K, Hannon GJ. Developmentally regulated piRNA clusters implicate MILI in transposon control. Science. 2007;316: 744–747. - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

