Homology Directed Knockin of Point Mutations in the Zebrafish tardbp and fus Genes in ALS Using the CRISPR/Cas9 System
- PMID: 26930076
- PMCID: PMC4773037
- DOI: 10.1371/journal.pone.0150188
Homology Directed Knockin of Point Mutations in the Zebrafish tardbp and fus Genes in ALS Using the CRISPR/Cas9 System
Abstract
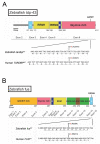
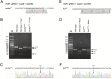
The methodology for site-directed editing of single nucleotides in the vertebrate genome is of considerable interest for research in biology and medicine. The clustered regularly interspaced short palindromic repeats (CRISPR)/CRISPR-associated protein 9 type II (Cas9) system has emerged as a simple and inexpensive tool for editing genomic loci of interest in a variety of animal models. In zebrafish, error-prone non-homologous end joining (NHEJ) has been used as a simple method to disrupt gene function. We sought to develop a method to easily create site-specific SNPs in the zebrafish genome. Here, we report simple methodologies for using CRISPR/Cas9-mediated homology directed repair using single-stranded oligodeoxynucleotide donor templates (ssODN) for site-directed single nucleotide editing, for the first time in two disease-related genes, tardbp and fus.
Conflict of interest statement
Figures


References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases
Research Materials
Miscellaneous

