Genome editing for crop improvement: Challenges and opportunities
- PMID: 26930114
- PMCID: PMC5033222
- DOI: 10.1080/21645698.2015.1129937
Genome editing for crop improvement: Challenges and opportunities
Abstract
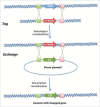
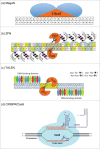
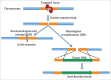
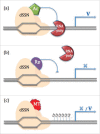
Genome or gene editing includes several new techniques to help scientists precisely modify genome sequences. The techniques also enables us to alter the regulation of gene expression patterns in a pre-determined region and facilitates novel insights into the functional genomics of an organism. Emergence of genome editing has brought considerable excitement especially among agricultural scientists because of its simplicity, precision and power as it offers new opportunities to develop improved crop varieties with clear-cut addition of valuable traits or removal of undesirable traits. Research is underway to improve crop varieties with higher yields, strengthen stress tolerance, disease and pest resistance, decrease input costs, and increase nutritional value. Genome editing encompasses a wide variety of tools using either a site-specific recombinase (SSR) or a site-specific nuclease (SSN) system. Both systems require recognition of a known sequence. The SSN system generates single or double strand DNA breaks and activates endogenous DNA repair pathways. SSR technology, such as Cre/loxP and Flp/FRT mediated systems, are able to knockdown or knock-in genes in the genome of eukaryotes, depending on the orientation of the specific sites (loxP, FLP, etc.) flanking the target site. There are 4 main classes of SSN developed to cleave genomic sequences, mega-nucleases (homing endonuclease), zinc finger nucleases (ZFNs), transcriptional activator-like effector nucleases (TALENs), and the CRISPR/Cas nuclease system (clustered regularly interspaced short palindromic repeat/CRISPR-associated protein). The recombinase mediated genome engineering depends on recombinase (sub-) family and target-site and induces high frequencies of homologous recombination. Improving crops with gene editing provides a range of options: by altering only a few nucleotides from billions found in the genomes of living cells, altering the full allele or by inserting a new gene in a targeted region of the genome. Due to its precision, gene editing is more precise than either conventional crop breeding methods or standard genetic engineering methods. Thus this technology is a very powerful tool that can be used toward securing the world's food supply. In addition to improving the nutritional value of crops, it is the most effective way to produce crops that can resist pests and thrive in tough climates. There are 3 types of modifications produced by genome editing; Type I includes altering a few nucleotides, Type II involves replacing an allele with a pre-existing one and Type III allows for the insertion of new gene(s) in predetermined regions in the genome. Because most genome-editing techniques can leave behind traces of DNA alterations evident in a small number of nucleotides, crops created through gene editing could avoid the stringent regulation procedures commonly associated with GM crop development. For this reason many scientists believe plants improved with the more precise gene editing techniques will be more acceptable to the public than transgenic plants. With genome editing comes the promise of new crops being developed more rapidly with a very low risk of off-target effects. It can be performed in any laboratory with any crop, even those that have complex genomes and are not easily bred using conventional methods.
Keywords: CRISPR/Cas; MegaN; SSN; SSR; ZFN and TALEN.
Figures

Similar articles
-
Emerging Genome Engineering Tools in Crop Research and Breeding.Methods Mol Biol. 2020;2072:165-181. doi: 10.1007/978-1-4939-9865-4_14. Methods Mol Biol. 2020. PMID: 31541446
-
An overview of genome engineering in plants, including its scope, technologies, progress and grand challenges.Funct Integr Genomics. 2023 Apr 6;23(2):119. doi: 10.1007/s10142-023-01036-w. Funct Integr Genomics. 2023. PMID: 37022538 Review.
-
Evolution in crop improvement approaches and future prospects of molecular markers to CRISPR/Cas9 system.Gene. 2020 Aug 30;753:144795. doi: 10.1016/j.gene.2020.144795. Epub 2020 May 22. Gene. 2020. PMID: 32450202 Review.
-
Modern Trends in Plant Genome Editing: An Inclusive Review of the CRISPR/Cas9 Toolbox.Int J Mol Sci. 2019 Aug 19;20(16):4045. doi: 10.3390/ijms20164045. Int J Mol Sci. 2019. PMID: 31430902 Free PMC article. Review.
-
Revisiting CRISPR/Cas-mediated crop improvement: Special focus on nutrition.J Biosci. 2020;45:137. J Biosci. 2020. PMID: 33361628
Cited by
-
Exploring the landscape of public attitudes towards gene-edited foods in Japan.Breed Sci. 2024 Mar;74(1):11-21. doi: 10.1270/jsbbs.23047. Epub 2024 Feb 22. Breed Sci. 2024. PMID: 39246435 Free PMC article.
-
CRISPR/Cas9 and Nanotechnology Pertinence in Agricultural Crop Refinement.Front Plant Sci. 2022 Apr 8;13:843575. doi: 10.3389/fpls.2022.843575. eCollection 2022. Front Plant Sci. 2022. PMID: 35463432 Free PMC article. Review.
-
CRISPR-Cas9: Tool for Qualitative and Quantitative Plant Genome Editing.Front Plant Sci. 2016 Nov 21;7:1740. doi: 10.3389/fpls.2016.01740. eCollection 2016. Front Plant Sci. 2016. PMID: 27917188 Free PMC article. Review.
-
Accelerating crop improvement via integration of transcriptome-based network biology and genome editing.Planta. 2025 Mar 17;261(4):92. doi: 10.1007/s00425-025-04666-5. Planta. 2025. PMID: 40095140 Review.
-
Breeding custom-designed crops for improved drought adaptation.Adv Genet (Hoboken). 2021 Sep 20;2(3):e202100017. doi: 10.1002/ggn2.202100017. eCollection 2021 Sep. Adv Genet (Hoboken). 2021. PMID: 36620433 Free PMC article. Review.
References
-
- Albert NW, Davies KM, Lewis DH, Zhang H, Montefiori M, Brendolise C, Boase MR, Ngo H, Jameson PE, Schwinn KE. A conserved network of transcriptional activators and repressors regulates anthocyanin pigmentation in eudicots. Plant cell 2014; 26:962-80; PMID:24642943; http://dx.doi.org/10.1105/tpc.113.122069 - DOI - PMC - PubMed
-
- Ali Z, Abul-Faraj A, Piatek M, Mahfouz MM. Activity and specificity of TRV-mediated gene editing in plants. Plant Signal Behav. 2015; 10(10):e1044191; PMID:26039254; http://dx.doi.org/10.1080/15592324.2015.1044191 - DOI - PMC - PubMed
-
- Allan AC, Hellens RP, Laing WA. MYB transcription factors that colour our fruit. Trends Plant Sci 2008; 13:99-102; PMID:18280199; http://dx.doi.org/10.1016/j.tplants.2007.11.012 - DOI - PubMed
-
- Allen BG, Weeks DL. Transgenic Xenopus laevis embryos can be generated using phiC31 integrase. Nat Methods 2005; 2:975-9; PMID:16299484; http://dx.doi.org/10.1038/nmeth814 - DOI - PMC - PubMed
-
- Allen BG, Weeks DL. Bacteriophage phiC31 integrase mediated transgenesis in Xenopus laevis for protein expression at endogenous levels. Methods Mol Biol 2009; 518:113-22; PMID:19085128; http://dx.doi.org/10.1007/978-1-59745-202-1_9 - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
