Biochemical reconstitution of TET1-TDG-BER-dependent active DNA demethylation reveals a highly coordinated mechanism
- PMID: 26932196
- PMCID: PMC4778062
- DOI: 10.1038/ncomms10806
Biochemical reconstitution of TET1-TDG-BER-dependent active DNA demethylation reveals a highly coordinated mechanism
Abstract
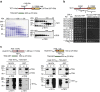
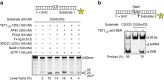
Cytosine methylation in CpG dinucleotides is an epigenetic DNA modification dynamically established and maintained by DNA methyltransferases and demethylases. Molecular mechanisms of active DNA demethylation began to surface only recently with the discovery of the 5-methylcytosine (5mC)-directed hydroxylase and base excision activities of ten-eleven translocation (TET) proteins and thymine DNA glycosylase (TDG). This implicated a pathway operating through oxidation of 5mC by TET proteins, which generates substrates for TDG-dependent base excision repair (BER) that then replaces 5mC with C. Yet, direct evidence for a productive coupling of TET with BER has never been presented. Here we show that TET1 and TDG physically interact to oxidize and excise 5mC, and proof by biochemical reconstitution that the TET-TDG-BER system is capable of productive DNA demethylation. We show that the mechanism assures a sequential demethylation of symmetrically methylated CpGs, thereby avoiding DNA double-strand break formation but contributing to the mutability of methylated CpGs.
Figures




References
-
- Bird A. DNA methylation patterns and epigenetic memory. Genes Dev. 16, 6–21 (2002). - PubMed
-
- Jones P. A. Functions of DNA methylation: islands, start sites, gene bodies and beyond. Nat. Rev. Genet. 13, 484–492 (2012). - PubMed
-
- Guo H. et al.. The DNA methylation landscape of human early embryos. Nature 511, 606–610 (2014). - PubMed
-
- Oswald J. et al.. Active demethylation of the paternal genome in the mouse zygote. Curr. Biol. 10, 475–478 (2000). - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

