Integrated metabolomics and metagenomics analysis of plasma and urine identified microbial metabolites associated with coronary heart disease
- PMID: 26932197
- PMCID: PMC4773756
- DOI: 10.1038/srep22525
Integrated metabolomics and metagenomics analysis of plasma and urine identified microbial metabolites associated with coronary heart disease
Abstract
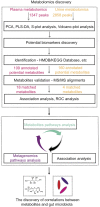
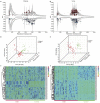
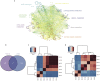
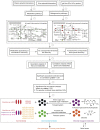
Coronary heart disease (CHD) is top risk factor for health in modern society, causing high mortality rate each year. However, there is no reliable way for early diagnosis and prevention of CHD so far. So study the mechanism of CHD and development of novel biomarkers is urgently needed. In this study, metabolomics and metagenomics technology are applied to discover new biomarkers from plasma and urine of 59 CHD patients and 43 healthy controls and trace their origin. We identify GlcNAc-6-P which has good diagnostic capability and can be used as potential biomarkers for CHD, together with mannitol and 15 plasma cholines. These identified metabolites show significant correlations with clinical biochemical indexes. Meanwhile, GlcNAc-6-P and mannitol are potential metabolites originated from intestinal microbiota. Association analysis on species and function levels between intestinal microbes and metabolites suggest a close correlation between Clostridium sp. HGF2 and GlcNAc-6-P, Clostridium sp. HGF2, Streptococcus sp. M143, Streptococcus sp. M334 and mannitol. These suggest the metabolic abnormality is significant and gut microbiota dysbiosis happens in CHD patients.
Figures

References
-
- Graninger D. J. et al.. Metabolomics in coronary heart disease. Heart. Metab 55, 8–12 (2012).
-
- Khan M. et al.. Contraindications and side effects of commonly used medications in coronary CT angiography. Int. J. Cardiovasc. Imaging 27, 441–449 (2011). - PubMed
-
- Huss J. M. et al.. Nuclear receptor signaling and cardiac energetics. Circ. Res 95, 568–578 (2004). - PubMed
-
- Chilton R. J. Pathophysiology of coronary heart disease: a brief review. JAOA 104, S5–S8 (2004). - PubMed
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Other Literature Sources

