Discovery of hantavirus circulating among Rattus rattus in French Mayotte island, Indian Ocean
- PMID: 26932442
- PMCID: PMC4851257
- DOI: 10.1099/jgv.0.000440
Discovery of hantavirus circulating among Rattus rattus in French Mayotte island, Indian Ocean
Abstract
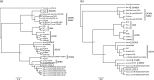
Hantaviruses are emerging zoonotic viruses that cause human diseases. In this study, sera from 642 mammals from La Réunion and Mayotte islands (Indian Ocean) were screened for the presence of hantaviruses by molecular analysis. None of the mammals from La Réunion island was positive, but hantavirus genomic RNA was discovered in 29/160 (18 %) Rattus rattus from Mayotte island. The nucleoprotein coding region was sequenced from the liver and spleen of all positive individuals allowing epidemiological and intra-strain variability analyses. Phylogenetic analysis based on complete coding genomic sequences showed that this Murinae-associated hantavirus is a new variant of Thailand virus. Further studies are needed to investigate hantaviruses in rodent hosts and in Haemorrhagic Fever with Renal Syndrome (HFRS) human cases.
Figures

References
-
- Cheke A., Hume J. (2008). Lost Land of the Dodo: The Ecological History of Mauritius, Réunion & Rodrigues London: T. & A.D. Poyser.
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases

