Selection of cell fate in the organ of Corti involves the integration of Hes/Hey signaling at the Atoh1 promoter
- PMID: 26932672
- PMCID: PMC4813338
- DOI: 10.1242/dev.129320
Selection of cell fate in the organ of Corti involves the integration of Hes/Hey signaling at the Atoh1 promoter
Abstract
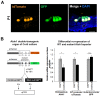
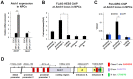
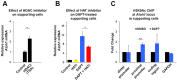
Determination of cell fate within the prosensory domain of the developing cochlear duct relies on the temporal and spatial regulation of the bHLH transcription factor Atoh1. Auditory hair cells and supporting cells arise in a wave of differentiation that patterns them into discrete rows mediated by Notch-dependent lateral inhibition. However, the mechanism responsible for selecting sensory cells from within the prosensory competence domain remains poorly understood. We show in mice that rather than being upregulated in rows of cells, Atoh1 is subject to transcriptional activation in groups of prosensory cells, and that highly conserved sites for Hes/Hey repressor binding in the Atoh1 promoter are needed to select the hair cell and supporting cell fate. During perinatal supporting cell transdifferentiation, which is a model of hair cell regeneration, we show that derepression is sufficient to induce Atoh1 expression, suggesting a mechanism for priming the 3' Atoh1 autoregulatory enhancer needed for hair cell expression.
Keywords: Atoh1; Cochlear development; Hair cell regeneration; Hes5; Mouse; Organ of Corti; Transdifferentiation.
© 2016. Published by The Company of Biologists Ltd.
Conflict of interest statement
The authors declare no competing or financial interests.
Figures




References
-
- Balasubramanyam K., Varier R. A., Altaf M., Swaminathan V., Siddappa N. B., Ranga U. and Kundu T. K. (2004). Curcumin, a novel p300/CREB-binding protein-specific inhibitor of acetyltransferase, represses the acetylation of histone/nonhistone proteins and histone acetyltransferase-dependent chromatin transcription. J. Biol. Chem. 279, 51163-51171. 10.1074/jbc.M409024200 - DOI - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

