Quantification provides a conceptual basis for convergent evolution
- PMID: 26932796
- PMCID: PMC6849873
- DOI: 10.1111/brv.12257
Quantification provides a conceptual basis for convergent evolution
Abstract
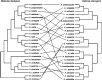
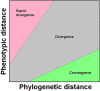
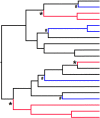
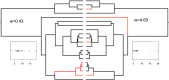
While much of evolutionary biology attempts to explain the processes of diversification, there is an important place for the study of phenotypic similarity across life forms. When similar phenotypes evolve independently in different lineages this is referred to as convergent evolution. Although long recognised, evolutionary convergence is receiving a resurgence of interest. This is in part because new genomic data sets allow detailed and tractable analysis of the genetic underpinnings of convergent phenotypes, and in part because of renewed recognition that convergence may reflect limitations in the diversification of life. In this review we propose that although convergent evolution itself does not require a new evolutionary framework, none the less there is room to generate a more systematic approach which will enable evaluation of the importance of convergent phenotypes in limiting the diversity of life's forms. We therefore propose that quantification of the frequency and strength of convergence, rather than simply identifying cases of convergence, should be considered central to its systematic comprehension. We provide a non-technical review of existing methods that could be used to measure evolutionary convergence, bringing together a wide range of methods. We then argue that quantification also requires clear specification of the level at which the phenotype is being considered, and argue that the most constrained examples of convergence show similarity both in function and in several layers of underlying form. Finally, we argue that the most important and impressive examples of convergence are those that pertain, in form and function, across a wide diversity of selective contexts as these persist in the likely presence of different selection pressures within the environment.
Keywords: convergence; evolutionary ecology; homoplasy; methods; parallelism; phylogenetic comparative methods.
© 2016 The Authors. Biological Reviews published by John Wiley & Sons Ltd on behalf of Cambridge Philosophical Society.
Figures

References
-
- Achaz, G. , Rodriguez‐Verdugo, A. , Gaut, B. S. & Tenaillon, O. (2014). The reproducibility of adaptation in the light of experimental evolution with whole genome sequencing. Ecological Genomics 781, 211–231. - PubMed
-
- Ackerly, D. D. & Donoghue, M. J. (1998). Leaf size, sapling allometry, and Corner's rules: phylogeny and correlated evolution in maples (Acer). The American Naturalist 152, 767–791. - PubMed
-
- Agrawal, A. A. & Fishbein, M. (2006). Plant defense syndromes. Ecology 87, S132–S149. - PubMed
-
- Arbuckle, K. , Bennett, C. M. & Speed, M. P. (2014). A simple measure of the strength of convergent evolution. Methods in Ecology and Evolution 5, 685–693.
-
- Archie, J. W. (1989). Homoplasy excess ratios: new indices for measuring levels of homoplasy in phylogenetic systematics and a critique of the consistency index. Systematic Biology 38, 253–269.
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Other Literature Sources

