The Combining Sites of Anti-lipid A Antibodies Reveal a Widely Utilized Motif Specific for Negatively Charged Groups
- PMID: 26933033
- PMCID: PMC4858963
- DOI: 10.1074/jbc.M115.712448
The Combining Sites of Anti-lipid A Antibodies Reveal a Widely Utilized Motif Specific for Negatively Charged Groups
Abstract
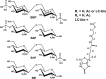
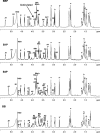
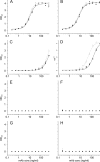
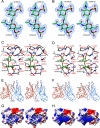
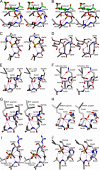
Lipopolysaccharide dispersed in the blood by Gram-negative bacteria can be a potent inducer of septic shock. One research focus has been based on antibody sequestration of lipid A (the endotoxic principle of LPS); however, none have been successfully developed into a clinical treatment. Comparison of a panel of anti-lipid A antibodies reveals highly specific antibodies produced through distinct germ line precursors. The structures of antigen-binding fragments for two homologous mAbs specific for lipid A, S55-3 and S55-5, have been determined both in complex with lipid A disaccharide backbone and unliganded. These high resolution structures reveal a conserved positively charged pocket formed within the complementarity determining region H2 loops that binds the terminal phosphates of lipid A. Significantly, this motif occurs in unrelated antibodies where it mediates binding to negatively charged moieties through a range of epitopes, including phosphorylated peptides used in diagnostics and therapeutics. S55-3 and S55-5 have combining sites distinct from anti-lipid A antibodies previously described (as a result of their separate germ line origin), which are nevertheless complementary both in shape and charge to the antigen. S55-3 and S55-5 display similar avidity toward lipid A despite possessing a number of different amino acid residues in their combining sites. Binding of lipid A occurs independent of the acyl chains, although the GlcN-O6 attachment point for the core oligosaccharide is buried in the combining site, which explains their inability to recognize LPS. Despite their lack of therapeutic potential, the observed motif may have significant immunological implications as a tool for engineering recombinant antibodies.
Keywords: X-ray crystallography; lipid A; lipopolysaccharide (LPS); monoclonal antibody; protein structure; recognition pocket.
© 2016 by The American Society for Biochemistry and Molecular Biology, Inc.
Figures

References
-
- Engel C., Brunkhorst F. M., Bone H. G., Brunkhorst R., Gerlach H., Grond S., Gruendling M., Huhle G., Jaschinski U., John S., Mayer K., Oppert M., Olthoff D., Quintel M., Ragaller M., et al. (2007) Epidemiology of sepsis in Germany: results from a national prospective multicenter study. Intensive Care Med. 33, 606–618 - PubMed
-
- Angus D. C., Linde-Zwirble W. T., Lidicker J., Clermont G., Carcillo J., and Pinsky M. R. (2001) Epidemiology of severe sepsis in the United States: analysis of incidence, outcome, and associated costs of care. Crit. Care Med. 29, 1303–1310 - PubMed
-
- Buttenschoen K., Radermacher P., and Bracht H. (2010) Endotoxin elimination in sepsis: physiology and therapeutic application. Langenbeck Arch. Surg. 395, 597–605 - PubMed
-
- Rietschel E. T., Brade H., Brade L., Brandenburg K., Schade U., Seydel U., Zähringer U., Galanos C., Lüderitz O., and Westphal O. (1987) Lipid A, the endotoxic center of bacterial lipopolysaccharides: relation of chemical structure to biological activity. Prog. Clin. Biol. Res. 231, 25–53 - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
LinkOut - more resources
Full Text Sources
Other Literature Sources

