Insight into infrageneric circumscription through complete chloroplast genome sequences of two Trillium species
- PMID: 26933149
- PMCID: PMC4823371
- DOI: 10.1093/aobpla/plw015
Insight into infrageneric circumscription through complete chloroplast genome sequences of two Trillium species
Abstract
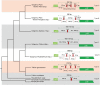
Genomic events including gene loss, duplication, pseudogenization and rearrangement in plant genomes are valuable sources for exploring and understanding the process of evolution in angiosperms. The family Melanthiaceae is distributed in temperate regions of the Northern Hemisphere and divided into five tribes (Heloniadeae, Chionographideae, Xerophylleae, Melanthieae and Parideae) based on the molecular phylogenetic analyses. At present, complete chloroplast genomes of the Melanthiaceae have been reported from three species. In the previous genomic study of Liliales, atrnI-CAU gene duplication event was reported fromParis verticillata, a member of Parideae. To clarify the significant genomic events of the tribe Parideae, we analysed the complete chloroplast genome sequences of twoTrilliumspecies representing two subgenera:TrilliumandPhyllantherum InTrillium tschonoskii(subgenusTrillium), the circular double-stranded cpDNA sequence of 156 852 bp consists of two inverted repeat (IR) regions of 26 501 bp each, a large single-copy (LSC) region of 83 981 bp and a small single-copy (SSC) region of 19 869 bp. The chloroplast genome sequence ofT. maculatum(subgenusPhyllantherum) is 157 359 bp in length, consisting of two IRs (25 535 bp), one SSC (19 949 bp) and one LSC (86 340 bp), and is longer than that ofT. tschonoskii The results showed that the cpDNAs of Parideae are highly conserved across genome structure, gene order and contents. However, the chloroplast genome ofT. maculatumcontained a 3.4-kb inverted sequence betweenndhCandrbcLin the LSC region, and it was a unique feature for subgeneraPhyllantherum In addition, we found three different types of gene duplication in the intergenic spacer betweenrpl23andycf2containingtrnI-CAU, which were in agreement with the circumscription of subgenera and sections in Parideae excludingT. govanianum These genomic features provide informative molecular markers for identifying the infrageneric taxa ofTrilliumand improve our understanding of the evolution patterns of Parideae in Melanthiaceae.
Keywords: Chloroplast genome; Trillium maculatum; Trillium tschonoskii, trnI-CAU; comparative genomics; gene duplication; single inversion.
Published by Oxford University Press on behalf of the Annals of Botany Company.
Figures




References
-
- Bodin SS, Kim JS, Kim J-H. 2013. Complete chloroplast genome of Chionographis japonica (Willd.) Maxim. (Melanthiaceae): comparative genomics and evaluation of universal primers for Liliales. Plant Molecular Biology Reporter 31:1407–1421. 10.1007/s11105-013-0616-x - DOI
-
- Bryan GJ, McNicoll J, Ramsay G, Meyer RC, De Jong WS. 1999. Polymorphic simple sequence repeat markers in chloroplast genomes of Solanaceous plants. Theoretical and Applied Genetics 99:859–867. 10.1007/s001220051306 - DOI
-
- Cai Z, Guisinger M, Kim H-G, Ruck E, Blazier JC, Mcmurtry V, Kuehl JV, Boore J, Jansen RK. 2008. Extensive reorganization of the plastid genome of Trifolium subterraneum (Fabaceae) is associated with numerous repeated sequences and novel DNA insertions. Journal of Molecular Evolution 67:696–704. 10.1007/s00239-008-9180-7 - DOI - PubMed
LinkOut - more resources
Full Text Sources
Other Literature Sources

