Nucleic Acid Modifications in Regulation of Gene Expression
- PMID: 26933737
- PMCID: PMC4779186
- DOI: 10.1016/j.chembiol.2015.11.007
Nucleic Acid Modifications in Regulation of Gene Expression
Abstract
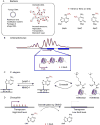
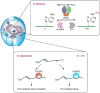
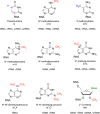
Nucleic acids carry a wide range of different chemical modifications. In contrast to previous views that these modifications are static and only play fine-tuning functions, recent research advances paint a much more dynamic picture. Nucleic acids carry diverse modifications and employ these chemical marks to exert essential or critical influences in a variety of cellular processes in eukaryotic organisms. This review covers several nucleic acid modifications that play important regulatory roles in biological systems, especially in regulation of gene expression: 5-methylcytosine (5mC) and its oxidative derivatives, and N(6)-methyladenine (6mA) in DNA; N(6)-methyladenosine (m(6)A), pseudouridine (Ψ), and 5-methylcytidine (m(5)C) in mRNA and long non-coding RNA. Modifications in other non-coding RNAs, such as tRNA, miRNA, and snRNA, are also briefly summarized. We provide brief historical perspective of the field, and highlight recent progress in identifying diverse nucleic acid modifications and exploring their functions in different organisms. Overall, we believe that work in this field will yield additional layers of both chemical and biological complexity as we continue to uncover functional consequences of known nucleic acid modifications and discover new ones.
Copyright © 2016 Elsevier Ltd. All rights reserved.
Figures

References
-
- Agris PF. The Importance of Being Modified: Roles of Modified Nucleosides and Mg2+ in RNA Structure and Function. In: Waldo EC, Klvle M, editors. Progress in Nucleic Acid Research and Molecular Biology. Academic Press; 1996. pp. 79–129. - PubMed
-
- Aran D, Toperoff G, Rosenberg M, Hellman A. Replication timing-related and gene body-specific methylation of active human genes. Hum Mol Gen. 2011;20:670–680. - PubMed
-
- Arnez JG, Steitz TA. Crystal Structure of Unmodified tRNAGln Complexed with Glutaminyl-tRNA Synthetase and ATP Suggests a Possible Role for Pseudo-Uridines in Stabilization of RNA Structure. Biochemistry. 1994;33:7560–7567. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

