EasyCloneMulti: A Set of Vectors for Simultaneous and Multiple Genomic Integrations in Saccharomyces cerevisiae
- PMID: 26934490
- PMCID: PMC4775045
- DOI: 10.1371/journal.pone.0150394
EasyCloneMulti: A Set of Vectors for Simultaneous and Multiple Genomic Integrations in Saccharomyces cerevisiae
Abstract
Saccharomyces cerevisiae is widely used in the biotechnology industry for production of ethanol, recombinant proteins, food ingredients and other chemicals. In order to generate highly producing and stable strains, genome integration of genes encoding metabolic pathway enzymes is the preferred option. However, integration of pathway genes in single or few copies, especially those encoding rate-controlling steps, is often not sufficient to sustain high metabolic fluxes. By exploiting the sequence diversity in the long terminal repeats (LTR) of Ty retrotransposons, we developed a new set of integrative vectors, EasyCloneMulti, that enables multiple and simultaneous integration of genes in S. cerevisiae. By creating vector backbones that combine consensus sequences that aim at targeting subsets of Ty sequences and a quickly degrading selective marker, integrations at multiple genomic loci and a range of expression levels were obtained, as assessed with the green fluorescent protein (GFP) reporter system. The EasyCloneMulti vector set was applied to balance the expression of the rate-controlling step in the β-alanine pathway for biosynthesis of 3-hydroxypropionic acid (3HP). The best 3HP producing clone, with 5.45 g.L(-1) of 3HP, produced 11 times more 3HP than the lowest producing clone, which demonstrates the capability of EasyCloneMulti vectors to impact metabolic pathway enzyme activity.
Conflict of interest statement
Figures
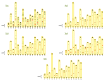


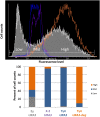
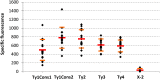
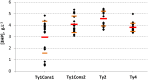
References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials

