FlyBase portals to human disease research using Drosophila models
- PMID: 26935103
- PMCID: PMC4826978
- DOI: 10.1242/dmm.023317
FlyBase portals to human disease research using Drosophila models
Abstract
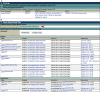
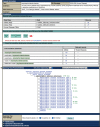
The use of Drosophila melanogaster as a model for studying human disease is well established, reflected by the steady increase in both the number and proportion of fly papers describing human disease models in recent years. In this article, we highlight recent efforts to improve the availability and accessibility of the disease model information in FlyBase (http://flybase.org), the model organism database for Drosophila. FlyBase has recently introduced Human Disease Model Reports, each of which presents background information on a specific disease, a tabulation of related disease subtypes, and summaries of experimental data and results using fruit flies. Integrated presentations of relevant data and reagents described in other sections of FlyBase are incorporated into these reports, which are specifically designed to be accessible to non-fly researchers in order to promote collaboration across model organism communities working in translational science. Another key component of disease model information in FlyBase is that data are collected in a consistent format --- using the evolving Disease Ontology (an open-source standardized ontology for human-disease-associated biomedical data) - to allow robust and intuitive searches. To facilitate this, FlyBase has developed a dedicated tool for querying and navigating relevant data, which include mutations that model a disease and any associated interacting modifiers. In this article, we describe how data related to fly models of human disease are presented in individual Gene Reports and in the Human Disease Model Reports. Finally, we discuss search strategies and new query tools that are available to access the disease model data in FlyBase.
Keywords: Disease model; Drosophila; FlyBase; Online resource.
© 2016. Published by The Company of Biologists Ltd.
Conflict of interest statement
The authors declare no competing or financial interests.
Figures

References
-
- Bellen H. J., Levis R. W., Liao G., He Y., Carlson J. W., Tsang G., Evans-Holm M., Hiesinger P. R., Schulze K. L., Rubin G. M. et al. (2004). The BDGP gene disruption project: single transposon insertions associated with 40% of Drosophila genes. Genetics 167, 761-781. 10.1534/genetics.104.026427 - DOI - PMC - PubMed
-
- Bellen H. J., Levis R. W., He Y., Carlson J. W., Evans-Holm M., Bae E., Kim J., Metaxakis A., Savakis C., Schulze K. L. et al. (2011). The Drosophila gene disruption project: progress using transposons with distinctive site specificities. Genetics 188, 731-743. 10.1534/genetics.111.126995 - DOI - PMC - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

