Bacterial Vegetative Insecticidal Proteins (Vip) from Entomopathogenic Bacteria
- PMID: 26935135
- PMCID: PMC4867366
- DOI: 10.1128/MMBR.00060-15
Bacterial Vegetative Insecticidal Proteins (Vip) from Entomopathogenic Bacteria
Erratum in
-
Correction for Chakroun et al., Bacterial Vegetative Insecticidal Proteins (Vip) from Entomopathogenic Bacteria.Microbiol Mol Biol Rev. 2016 Aug 10;80(3):iii. doi: 10.1128/MMBR.00039-16. Print 2016 Sep. Microbiol Mol Biol Rev. 2016. PMID: 27512101 Free PMC article. No abstract available.
Abstract
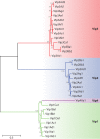
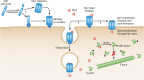
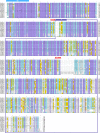
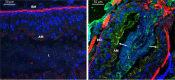
Entomopathogenic bacteria produce insecticidal proteins that accumulate in inclusion bodies or parasporal crystals (such as the Cry and Cyt proteins) as well as insecticidal proteins that are secreted into the culture medium. Among the latter are the Vip proteins, which are divided into four families according to their amino acid identity. The Vip1 and Vip2 proteins act as binary toxins and are toxic to some members of the Coleoptera and Hemiptera. The Vip1 component is thought to bind to receptors in the membrane of the insect midgut, and the Vip2 component enters the cell, where it displays its ADP-ribosyltransferase activity against actin, preventing microfilament formation. Vip3 has no sequence similarity to Vip1 or Vip2 and is toxic to a wide variety of members of the Lepidoptera. Its mode of action has been shown to resemble that of the Cry proteins in terms of proteolytic activation, binding to the midgut epithelial membrane, and pore formation, although Vip3A proteins do not share binding sites with Cry proteins. The latter property makes them good candidates to be combined with Cry proteins in transgenic plants (Bacillus thuringiensis-treated crops [Bt crops]) to prevent or delay insect resistance and to broaden the insecticidal spectrum. There are commercially grown varieties of Bt cotton and Bt maize that express the Vip3Aa protein in combination with Cry proteins. For the most recently reported Vip4 family, no target insects have been found yet.
Copyright © 2016, American Society for Microbiology. All Rights Reserved.
Figures












References
-
- Sanchis V. 2011. From microbial sprays to insect-resistant transgenic plants: history of the biopesticide Bacillus thuringiensis. A review. Agron Sustain Dev 31:217–231. doi: 10.1051/agro/2010027. - DOI
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

