A bead-based cleavage method for large-scale identification of protease substrates
- PMID: 26935269
- PMCID: PMC4776233
- DOI: 10.1038/srep22645
A bead-based cleavage method for large-scale identification of protease substrates
Abstract
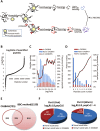
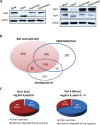
Proteolysis is a major form of post translational modification which occurs when a protease cleaves peptide bonds in a target protein to modify its activity. Tracking protease substrates is indispensable for understanding its cellular functions. However, it is difficult to directly identify protease substrates because the end products of proteolysis, the cleaved protein fragments, must be identified among the pool of cellular proteins. Here we present a bead-based cleavage approach using immobilized proteome as the screening library to identify protease substrates. This method enables efficient separation of proteolyzed proteins from background protein mixture. Using caspase-3 as the model protease, we have identified 1159 high confident substrates, among which, strikingly, 43.9% of substrates undergo degradation during apoptosis. The huge number of substrates and positive support of in vivo evidence indicate that the BBC method is a powerful tool for protease substrates identification.
Figures
Similar articles
-
Cell-based identification of natural substrates and cleavage sites for extracellular proteases by SILAC proteomics.Methods Mol Biol. 2009;539:131-53. doi: 10.1007/978-1-60327-003-8_8. Methods Mol Biol. 2009. PMID: 19377966
-
A Novel 2-DE-Based Proteomic Analysis to Identify Multiple Substrates for Specific Protease in Neuronal Cells.Methods Mol Biol. 2017;1598:229-245. doi: 10.1007/978-1-4939-6952-4_10. Methods Mol Biol. 2017. PMID: 28508364
-
Proteolysis to Identify Protease Substrates: Cleave to Decipher.Proteomics. 2018 Jul;18(13):e1800011. doi: 10.1002/pmic.201800011. Epub 2018 Jun 10. Proteomics. 2018. PMID: 29710386 Review.
-
Deep profiling of protease substrate specificity enabled by dual random and scanned human proteome substrate phage libraries.Proc Natl Acad Sci U S A. 2020 Oct 13;117(41):25464-25475. doi: 10.1073/pnas.2009279117. Epub 2020 Sep 24. Proc Natl Acad Sci U S A. 2020. PMID: 32973096 Free PMC article.
-
Methods for the proteomic identification of protease substrates.Curr Opin Chem Biol. 2009 Dec;13(5-6):503-9. doi: 10.1016/j.cbpa.2009.07.026. Epub 2009 Sep 2. Curr Opin Chem Biol. 2009. PMID: 19729334 Free PMC article. Review.
Cited by
-
Caspases maintain tissue integrity by an apoptosis-independent inhibition of cell migration and invasion.Nat Commun. 2018 Jul 18;9(1):2806. doi: 10.1038/s41467-018-05204-6. Nat Commun. 2018. PMID: 30022065 Free PMC article.
-
Alterations in the nucleocytoplasmic transport in apoptosis: Caspases lead the way.Cell Prolif. 2018 Oct;51(5):e12467. doi: 10.1111/cpr.12467. Epub 2018 Jun 26. Cell Prolif. 2018. PMID: 29947118 Free PMC article. Review.
-
Protease Activity Profiling of Snake Venoms Using High-Throughput Peptide Screening.Toxins (Basel). 2019 Mar 19;11(3):170. doi: 10.3390/toxins11030170. Toxins (Basel). 2019. PMID: 30893860 Free PMC article.
-
A global view of the human post-translational modification landscape.Biochem J. 2023 Aug 30;480(16):1241-1265. doi: 10.1042/BCJ20220251. Biochem J. 2023. PMID: 37610048 Free PMC article. Review.
References
-
- Puente X. S., Sanchez L. M., Overall C. M. & Lopez-Otin C. Human and mouse proteases: a comparative genomic approach. Nat Rev Genet. 4, 544–558 (2003). - PubMed
-
- Doucet A., Butler G. S., Rodriguez D., Prudova A. & Overall C. M. Metadegradomics: toward in vivo quantitative degradomics of proteolytic post-translational modifications of the cancer proteome. Mol Cell Proteomics. 7, 1925–1951 (2008). - PubMed
-
- Dewachter I. & Van Leuven F. Secretases as targets for the treatment of Alzheimer’s disease: the prospects. Lancet Neurol. 1, 409–416 (2002). - PubMed
-
- Turk B. Targeting proteases: successes, failures and future prospects. Nat Rev Drug Discov. 5, 785–799 (2006). - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

