Lrig1 is a cell-intrinsic modulator of hippocampal dendrite complexity and BDNF signaling
- PMID: 26935556
- PMCID: PMC4818773
- DOI: 10.15252/embr.201541218
Lrig1 is a cell-intrinsic modulator of hippocampal dendrite complexity and BDNF signaling
Abstract
Even though many extracellular factors have been identified as promoters of general dendritic growth and branching, little is known about the cell-intrinsic modulators that allow neurons to sculpt distinctive patterns of dendrite arborization. Here, we identify Lrig1, a nervous system-enriched LRR protein, as a key physiological regulator of dendrite complexity of hippocampal pyramidal neurons. Lrig1-deficient mice display morphological changes in proximal dendrite arborization and defects in social interaction. Specifically, knockdown of Lrig1 enhances both primary dendrite formation and proximal dendritic branching of hippocampal neurons, two phenotypes that resemble the effect of BDNF on these neurons. In addition, we show that Lrig1 physically interacts with TrkB and attenuates BDNF signaling. Gain and loss of function assays indicate that Lrig1 restricts BDNF-induced dendrite morphology. Together, our findings reveal a novel and essential role of Lrig1 in regulating morphogenic events that shape the hippocampal circuits and establish that the assembly of TrkB with Lrig1 represents a key mechanism for understanding how specific neuronal populations expand the repertoire of responses to BDNF during brain development.
Keywords: Lrig1; TrkB; dendrite morphogenesis; hippocampal neurons; neurotrophins.
© 2016 The Authors.
Figures
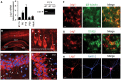
- A
Quantitative analysis of developmental expression of Lrig1 mRNA in rat hippocampus by real‐time RT–PCR. The results are shown as mean ± SEM of n = 3 independent assays. The levels of Lrig1 mRNA were normalized using the expression of the housekeeping gene Tbp (TATA box‐binding protein). The insert shows the expression of Lrig1 in embryonic E17.5 rat hippocampus examined by RT–PCR. Control sample without reverse transcriptase (‐RT) is also shown.
- B
Localization of Lrig1 (red) in coronal sections from P15 rat brain by immunofluorescence. Anti‐Lrig1ECD antibodies label dentate gyrus cells, CA1–CA3 hippocampal neurons, and pyramidal cortical neurons (asterisk). Scale bar, 400 μm.
- C–E
Confocal image of cortical (C) and hippocampal (D, E) pyramidal neurons stained with rabbit anti‐Lrig1ECD antibody. Arrows indicate Lrig1 staining in proximal segments of apical dendrites of CA1–CA3 hippocampal and pyramidal cortical neurons (layer V). Scale bars, 20 μm.
- F
Immunofluorescence staining of Lrig1 (red) with anti‐Lrig1ECD antibody and the neuronal marker βIII‐tubulin (green) in dissociated hippocampal cells cultured for 7 DIV. Yellow indicates neuronal expression of Lrig1. Scale bar, 15 μm.
- G
Immunofluorescence staining of Lrig1 (red) with anti‐Lrig1ECD antibody and the astrocytic marker S100β (green) in dissociated hippocampal cells cultured for 7 DIV. Scale bar, 20 μm.
- H
Localization of Lrig1 (red) with anti‐Lrig1ECD antibody and the somatodendritic marker MAP‐2 (blue) by immunocytochemistry in dissociated rat hippocampal neurons after 12 DIV. Scale bar, 20 μm.
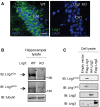
Confocal images of CA1 hippocampal pyramidal neurons labeled for Lrig1 (green). The specificity of the rabbit polyclonal antibody anti‐Lrig1 extracellular domain (gift from Dr. S. Itami) was additionally tested by immunofluorescence labeling of P15 brain sections from wild‐type and Lrig1‐null mice. Scale bar, 20 μm.
Confirmation of Lrig1 ablation by immunoblot (IB) analysis of hippocampal homogenates obtained from wild‐type and Lrig1‐knockout mice probed with antibodies against Lrig1 and tubulin.
Antibody specificity was confirmed by immunoblotting of COS cell extracts transfected to ectopically express Flag‐Lrig1, HA‐Lrig2, or Flag‐Lrig3. ICD and ECD indicate intracellular and extracellular domains, respectively.
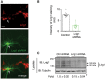
Lrig1 knockdown efficiency was additionally tested by immunofluorescence staining of mouse primary hippocampal neurons transfected with Lrig1‐shRNA‐GFP vector to knockdown endogenous Lrig1 expression. Lrig1 expression was specifically abrogated in primary hippocampal neurons transfected with Lrig1‐shRNA but not in untransfected neighboring cells. Arrow shows a representative Lrig1‐shRNA‐GFP‐transfected neuron negative for Lrig1 staining. Note the prominent proximal branching observed after Lrig1 downregulation. Scale bar, 20 μm.
The histogram shows the quantification of endogenous Lrig1 immunostaining in GFP‐positive hippocampal neurons transfected with either control or Lrig1‐shRNA constructs. The results are presented as individual values and means of a representative assay. n = 2 independent experiments were performed.
Endogenous levels of Lrig1 protein were analyzed by immunoblot in MN1 cells transfected with scrambled (Ctrl) or Lrig1‐shRNA vectors. Numbers below the lanes indicate fold changes relative to control cells normalized to the levels of β‐tubulin. Values are presented as averages ± SD of n = 3 independent assays. *P < 0.05 (Student's t‐test).
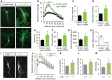
- A
Representative images of mouse hippocampal neurons transfected with either GFP‐expressing control or Lrig1‐shRNA vector at 9 DIV and maintained for 3 additional days in vitro (9 + 3 DIV). Scale bar, 15 μm. Boxed area represents a higher magnification image showing the profuse proximal dendritic arborization of Lrig1‐shRNA‐transfected neurons.
- B
Sholl analysis of the dendritic arbor from hippocampal neurons transfected with either control or Lrig1‐shRNA‐GFP vector at 9 DIV and maintained for 3 additional days in vitro (9 + 3 DIV). Data are shown as mean ± SEM of n = 3 independent experiments. *P < 0.05 and **P < 0.01 by two‐way ANOVA followed by Bonferroni multiple comparisons test.
- C–G
Quantification of primary (C) and secondary (D) dendrites as well as total dendritic branching (E), terminal dendritic points (F), and total dendritic length (G) of hippocampal neurons transfected with either control or Lrig1‐shRNA‐GFP vector. The results are shown as mean ± SEM of n = 3 independent experiments. *P < 0.05 by Student's t‐test.
- H
Knockdown efficiency was analyzed by real‐time RT–PCR in MN1 cells transfected with control or Lrig1‐shRNA vectors. Transfected cells were enriched by puromycin treatment in order to increase the population of cells expressing control or Lrig1‐shRNA constructs. Data are shown as individual values of a representative assay measured in triplicates. n = 2 independent experiments were performed.
- I
Representative images of MAP‐2 immunostained hippocampal neurons obtained from wild‐type and Lrig1‐deficient mice cultured for 7 days in vitro (7 DIV). Scale bar, 15 μm.
- J
Sholl analysis of the dendritic arbor from MAP‐2 stained hippocampal neurons (7 DIV) isolated from wild‐type and Lrig1‐deficient mice. Data are shown as mean ± SEM of n = 3 independent experiments. *P < 0.05 by two‐way ANOVA followed by Bonferroni multiple comparisons test.
- K–M
Quantification of the number of primary dendrites (K), secondary dendrites (L), and total dendritic branching (M) from MAP‐2 stained hippocampal neurons (7 DIV) isolated from wild‐type and Lrig1‐deficient mice. The results are shown as mean ± SEM of n = 3 independent experiments. *P < 0.05 by Student's t‐test.
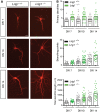
Representative images of MAP‐2‐immunostained hippocampal neurons obtained from wild‐type and Lrig1‐deficient mice and cultured for 7, 10, and 14 days in vitro (DIV). Scale bar, 20 μm.
Quantification of the number of primary dendrites, dendritic branching, and total dendritic length of MAP‐2‐stained hippocampal neurons isolated from wild‐type and Lrig1‐deficient mice and cultured for 7, 10, and 14 DIV. Results are shown as individual data points of a representative experiment. A total number of 30 neurons were analyzed per condition. Similar results were obtained in n = 3 independent assays.
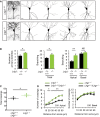
Representative images and drawings of Golgi‐stained hippocampal CA1 pyramidal neurons from 4‐week‐old wild‐type and Lrig1‐null littermate mice. Scale bar, 15 μm.
Quantification of the number of primary dendrites and branching of apical and basal dendritic arbors of hippocampal CA1 pyramidal neurons from 4‐ to 5‐week‐old control (wild‐type/heterozygous) and Lrig1‐null littermate mice. The results are shown as mean ± SEM of independent determinations performed in n = 4 mice of each genotype. *P < 0.05, **P < 0.001 by Student's t‐test. NS, not significant.
Cumulative dendrite crossings of concentric circles of increasing radius (10 μm ring interval) centering the reference point at the cell body. These values were obtained by Sholl analysis and represent the summatory of the dendritic crossings registered within the first 60 μm closest to the soma. The results are shown as mean ± SEM of independent determinations performed in n = 4 mice of each genotype. *P < 0.05 by Student's t‐test.
Sholl analysis of apical and basal dendritic arbors of hippocampal CA1 pyramidal neurons from 4‐ to 5‐week‐old control (wild‐type/heterozygous) and Lrig1‐null littermate mice. The results are shown as mean ± SEM. *P < 0.05, **P < 0.001 by two‐way ANOVA followed by Bonferroni multiple comparisons test.
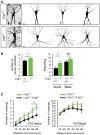
Representative images and drawings of Golgi‐stained hippocampal CA3 pyramidal neurons from 4‐week‐old wild‐type and Lrig1‐null littermate mice. Scale bar, 15 μm.
Quantification of the number of primary dendrites and proximal dendritic branching of apical and basal dendritic arbors of hippocampal CA3 pyramidal neurons from 4‐ to 5‐week‐old control (wild type/heterozygous) and Lrig1‐null littermate mice.
Sholl analysis of apical and basal dendritic arbors of hippocampal CA3 pyramidal neurons from 4‐ to 5‐week‐old control (wild type/heterozygous) and Lrig1‐null littermate mice.
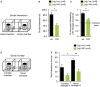
- A
Schematic diagram of the social interaction device indicating the social and the empty chambers.
- B, C
Mice were simultaneously exposed to an empty container and a caged unfamiliar juvenile mouse (social enclosure, stranger 1). Lrig1‐mutant mice exhibit social interaction defects as determined by the time spent interacting (sniffing) with the stranger enclosure (B) and the percentage of total interaction time with stranger in the three‐chamber social interaction test (C). Dashed line in (C) represents chance‐level performance (i.e., 50%) when mice equally explore the social enclosure and the empty container. Data represent means ± SEM of independent determinations performed in n = 8–9 mice of each genotype, and the statistical significance between wt and knockout mice is *P < 0.05 by Student's t‐test.
- D
Schematic diagram of the social novelty test. In the test for social novelty, a second stranger (stranger 2) mouse was introduced in the empty container.
- E
In the preference for social novelty task, wt mice showed a preference for social novelty, while mutant mice showed no significant preference for novel target (stranger 2). Mutants also spent significantly less time interacting with the novel mice compared to controls. Data represent means ± SEM of independent determinations performed in n = 8–9 mice of each genotype, and the statistical significance is as follows: *P < 0.05 and **P < 0.005 by ANOVA followed by Student–Newman–Keuls' multiple comparisons test.
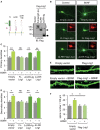
- A
Schematic representation of Lrig1 mutants is shown on the left. Expression levels of these mutants were analyzed in transfected cell extracts by immunoblotting with anti‐Flag antibodies.
- B
Representative images of rat hippocampal neurons transfected at DIV8 with empty vector, full‐length (FL) Flag‐tagged Lrig1, or Lrig1 mutant lacking the LRR domain (ΔLRR) in combination with an enhanced green fluorescent protein (GFP) expression vector. After transfection at 9 DIV, neurons were cultured in the absence or in the presence of BDNF (30 ng/ml) for 48 h. Then, hippocampal cultures at 11 DIV were fixed and stained with anti‐Flag antibodies to control Lrig1 expression. Scale bar, 15 μm.
- C, D
Quantification of the effects of Flag‐Lrig1 constructs on BDNF‐induced primary (C) and secondary (D) dendrite formation of hippocampal neurons treated as indicated in (A). The results are shown as mean ± SEM of 3 independent experiments. *P < 0.05, **P < 0.01 by one‐way ANOVA followed by Tukey's multiple comparisons test. NS, not significant.
- E
Representative confocal images of dendritic shafts containing spines from hippocampal neurons transfected at 15 DIV with either control vector or Flag‐Lrig1 construct together with GFP. After transfection, neurons were cultured in the absence or in the presence of BDNF (30 ng/ml) for 48 h (15 + 3 DIV). Scale bar, 5 μm.
- F
Quantification of the effect of Lrig1 overexpression on neurotrophin‐induced spine density. The results are shown as mean ± SEM of n = 3 independent experiments. *P < 0.05 by one‐way ANOVA followed by Student–Newman–Keuls' multiple comparisons test.

- A
Coexpression of Lrig1 (red) and TrkB (green) in primary rat hippocampal neurons. Boxed area represents a higher magnification image showing a high colocalization between TrkB and Lrig1 in a pyramidal neuron. Yellow indicates regions of colocalization. Scale bar, 30 μm. Data represent n = 3 independent experiments.
- B
Quantitative analysis of Lrig1 mRNA expression by real‐time RT–PCR. Rat hippocampal cultures (10 DIV) were treated with BDNF (50 ng/ml) during the indicated times. The levels were normalized using the expression of the housekeeping gene Tbp. The results are shown as mean ± SD of n = 3 independent experiments. *P < 0.05 vs. control group by one‐way ANOVA followed by Dunnett's test.
- C, D
Coimmunoprecipitation between Flag‐Lrig1 and HA‐TrkB (C) or between Flag‐Lrig3 and HA‐TrkB (D) overexpressed in HEK293 cells. Cell extracts were analyzed by immunoprecipitation with anti‐Flag antibodies followed by immunoblot (IB) with antibodies against HA. Reprobing of the same blots with anti‐Flag antibodies is shown below. The bottom panel shows HA expression in total lysates. Data represent n = 3 independent experiments.
- E
Coimmunoprecipitation between HA‐TrkB and Flag‐Lrig1 or between HA‐TrkB and HA‐Lrig2 exogenously expressed in HEK293 cells. Cell extracts were analyzed by IP with anti‐pan‐Trk antibodies followed by IB with antibodies against Flag or Lrig2. Reprobing of the same blots with anti‐TrkB antibodies is shown below. The bottom panels show Flag and Lrig2 expression in total lysates. Data represent n = 2 independent assays.
- F
In vivo interaction between Lrig1 and TrkB. Coimmunoprecipitation between Lrig1 and TrkB receptor endogenously expressed from P15 rat hippocampal tissue extracts. Samples were equally divided into two parts, and then, the analysis was done by immunoprecipitation with control (anti‐HA epitope tag antibody) or anti‐Lrig1 antibodies, followed by immunoblotting with anti‐TrkB antibody. Reprobing of the same blot with anti‐Lrig1 antibody is also shown. Expression of TrkB in one aliquot of the starting material (indicated as lysate) is included. Arrow indicates the band of TrkB coimmunoprecipitated with anti‐Lrig1 antibody. Similar results were obtained in n = 3 independent assays.
- G
Ligand‐dependent activation of TrkB (p‐TrkB) was evaluated by transient transfection of HA‐TrkB plasmid with either a control or a Flag‐Lrig1 vector into HEK cells. After 36 h, cells were serum‐starved and stimulated with or without BDNF (30 ng/ml) for 15 min. The level of TrkB activation (p‐TrkB) was evaluated in total cell lysates by immunoblotting (IB) with a specific antibody that recognizes TrkB phosphorylated in tyrosine 705 (pY705). Reprobing of the same blot with anti‐HA and anti‐Flag antibodies is shown. Fold of p‐TrkB change relative to total TrkB is indicated. Similar results were obtained in n = 3 independent assays.
- H
TrkB ubiquitination was evaluated by transient transfection of HA‐TrkB plasmid with either a control or a Flag‐Lrig1 vector into MN1 cells. After 36 h, cells were serum‐starved, pre‐treated with the cell‐permeable proteasome inhibitor MG‐132 (20 μM), and stimulated with BDNF for 15 min. Total lysates were immunoprecipitated with anti‐HA antibodies followed by immunoblot (IB) with antibodies against ubiquitin. Reprobing of the same blot with anti‐HA antibodies is also shown. TrkB activation (p‐TrkB) was evaluated in cell lysates. Reprobing of the same blot with anti‐TrkB and anti‐Flag antibodies is also shown. Fold of p‐TrkB (p‐Y705) change relative to total TrkB is indicated. Data represent n = 3 independent assays.
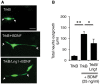
Representative images of PC12 cells expressing GFP and HA‐TrkB in the presence or in the absence of Flag‐Lrig1. Cells were fixed 72 h after stimulation with BDNF. Arrows indicate neurite tips. Scale bar, 50 μm.
Quantification of total neurite outgrowth expressed in μm. The results are shown as mean ± SEM of n = 3 independent experiments. *P < 0.05 and **P < 0.01 by ANOVA followed by Bonferroni test.
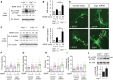
- A
Immunoblot showing TrkB activation in hippocampal neurons cultured from Lrig1 heterozygous (+/−) and Lrig1 knockout (−/−) mice littermates treated in the absence or in the presence of BDNF (30 ng/ml) for 30 min. Reprobing of the same blot with anti‐βIII‐tubulin is shown as a loading control.
- B
Fold of TrkB activation relative to unstimulated control group (phospho‐TrkB at Tyr705) in hippocampal neurons cultured from Lrig1 (+/+; +/−) and Lrig1 (−/−) mice treated in the absence or in the presence of BDNF (30 ng/ml) for 30 min. Results are presented as mean ± SEM of n = 4 independent experiments (*P < 0.05 by Student's t‐test).
- C
Immunoblot showing MAPK activation in hippocampal neurons cultured from Lrig1 wild‐type (+/+) and Lrig1‐deficient (−/−) mice littermates treated in the absence or in the presence of BDNF (30 ng/ml) for the indicated times. Reprobing of the same blot with anti‐βIII‐tubulin is shown as a loading control. Fold of MAPK activation relative to tubulin is indicated.
- D
Fold of MAPK activation (P‐MAPK) relative to untreated control group in hippocampal neurons cultured from Lrig1 (+/+; +/−) and Lrig1 (−/−) mice treated in the absence or in the presence of BDNF (30 ng/ml) for 30 min. Results are presented as mean ± SEM of n = 4 independent experiments (*P < 0.05 by Student's t‐test).
- E
Representative images of mouse hippocampal neurons transfected at 4 DIV with either GFP‐expressing control vector or Lrig1‐shRNA‐GFP plasmid. After transfection, neurons were cultured in the absence or in the presence of BDNF (25 ng/ml) for 48 h (7 DIV). Arrows indicate branching points along the principal dendrite. Scale bar, 10 μm.
- F–I
Quantification of primary dendrites (F), secondary dendrites (G), total branching (H), and branching of the principal dendrite (I) of hippocampal neurons treated as indicated in (E). The results are shown as individual values and the means of a representative assay. A total of 30 neurons were analyzed per experimental condition. Similar results were obtained in n = 3 independent experiments.
- J
Lrig1 +/+ and Lrig1 −/− hippocampal lysates from postnatal day 11 (P11) mice were immunoblotted against phospho‐TrkB (p‐TrkB) at Tyr705 and total TrkB. Quantification (mean ± SD) of the p‐TrkB/TrkB ratio between Lrig1 +/+ (n = 4) and Lrig1 −/− (n = 3) hippocampi is also shown.
References
-
- Parrish JZ, Emoto K, Kim MD, Jan YN (2007) Mechanisms that regulate establishment, maintenance, and remodeling of dendritic fields. Annu Rev Neurosci 30: 399–423 - PubMed
-
- Wong RO, Ghosh A (2002) Activity‐dependent regulation of dendritic growth and patterning. Nat Rev Neurosci 3: 803–812 - PubMed
-
- Whitford KL, Dijkhuizen P, Polleux F, Ghosh A (2002) Molecular control of cortical dendrite development. Annu Rev Neurosci 25: 127–149 - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous

