Itk is required for Th9 differentiation via TCR-mediated induction of IL-2 and IRF4
- PMID: 26936133
- PMCID: PMC4782063
- DOI: 10.1038/ncomms10857
Itk is required for Th9 differentiation via TCR-mediated induction of IL-2 and IRF4
Abstract
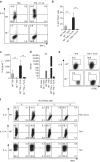
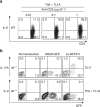
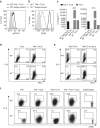
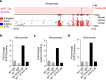
Th9 cells produce interleukin (IL)-9, a cytokine implicated in allergic asthma and autoimmunity. Here we show that Itk, a mediator of T cell receptor signalling required for Th2 immune responses and the development of asthma, is a positive regulator of Th9 differentiation. In a model of allergic lung disease, Itk-deficient mice show reduced pulmonary inflammation and IL-9 production by T cells and innate lymphoid type 2 cells (ILC2), despite normal early induction of ILC2s. In vitro, Itk(-/-) CD4(+) T cells do not produce IL-9 and have reduced levels of IRF4 (Interferon Regulator Factor 4), a critical transcription factor for effector T cell function. Both IL-9 and IRF4 expression are rescued by either IL-2 or constitutively active STAT5, but not NFATc1. STAT5 binds the Irf4 promoter, demonstrating one mechanism by which IL-2 rescues weakly activated T cells. Itk inhibition also reduces IL-9 expression by human T cells, implicating ITK as a key regulator of Th9 induction.
Figures





References
-
- Schmitt E., Klein M. & Bopp T. Th9 cells, new players in adaptive immunity. Trends Immunol. 35, 61–68 (2014). - PubMed
-
- Erpenbeck V. J. et al.. Segmental allergen challenge in patients with atopic asthma leads to increased IL-9 expression in bronchoalveolar lavage fluid lymphocytes. J. Allergy Clin. Immunol. 111, 1319–1327 (2003). - PubMed
-
- Hoppenot D. et al.. Peripheral blood Th9 cells and eosinophil apoptosis in asthma patients. Medicina (Kaunas) 51, 10–17 (2015). - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials
Miscellaneous

