Stimulation of translation by human Unr requires cold shock domains 2 and 4, and correlates with poly(A) binding protein interaction
- PMID: 26936655
- PMCID: PMC4776140
- DOI: 10.1038/srep22461
Stimulation of translation by human Unr requires cold shock domains 2 and 4, and correlates with poly(A) binding protein interaction
Abstract
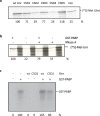
The RNA binding protein Unr, which contains five cold shock domains, has several specific roles in post-transcriptional control of gene expression. It can act as an activator or inhibitor of translation initiation, promote mRNA turnover, or stabilise mRNA. Its role depends on the mRNA and other proteins to which it binds, which includes cytoplasmic poly(A) binding protein 1 (PABP1). Since PABP1 binds to all polyadenylated mRNAs, and is involved in translation initiation by interaction with eukaryotic translation initiation factor 4G (eIF4G), we investigated whether Unr has a general role in translational control. We found that Unr strongly stimulates translation in vitro, and mutation of cold shock domains 2 or 4 inhibited its translation activity. The ability of Unr and its mutants to stimulate translation correlated with its ability to bind RNA, and to interact with PABP1. We found that Unr stimulated the binding of PABP1 to mRNA, and that Unr was required for the stable interaction of PABP1 and eIF4G in cells. siRNA-mediated knockdown of Unr reduced the overall level of cellular translation in cells, as well as that of cap-dependent and IRES-dependent reporters. These data describe a novel role for Unr in regulating cellular gene expression.
Figures



References
-
- Brown E. C. & Jackson R. J. All five cold-shock domains of unr (upstream of N-ras) are required for stimulation of human rhinovirus RNA translation. J Gen Virol 85, 2279–2287 (2004). - PubMed
-
- Anderson E. C., Hunt S. L. & Jackson R. J. Internal initiation of translation from the human rhinovirus-2 internal ribosome entry site requires the binding of Unr to two distinct sites on the 5′ untranslated region. J Gen Virol 88, 3043–3052 (2007). - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
Miscellaneous

