Motif mediated protein-protein interactions as drug targets
- PMID: 26936767
- PMCID: PMC4776425
- DOI: 10.1186/s12964-016-0131-4
Motif mediated protein-protein interactions as drug targets
Abstract
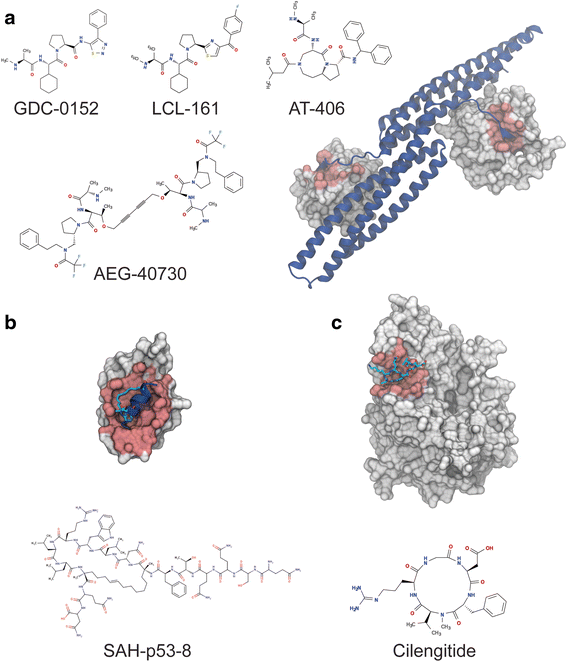
Protein-protein interactions (PPI) are involved in virtually every cellular process and thus represent an attractive target for therapeutic interventions. A significant number of protein interactions are frequently formed between globular domains and short linear peptide motifs (DMI). Targeting these DMIs has proven challenging and classical approaches to inhibiting such interactions with small molecules have had limited success. However, recent new approaches have led to the discovery of potent inhibitors, some of them, such as Obatoclax, ABT-199, AEG-40826 and SAH-p53-8 are likely to become approved drugs. These novel inhibitors belong to a wide range of different molecule classes, ranging from small molecules to peptidomimetics and biologicals. This article reviews the main reasons for limited success in targeting PPIs, discusses how successful approaches overcome these obstacles to discovery promising inhibitors for human protein double minute 2 (HDM2), B-cell lymphoma 2 (Bcl-2), X-linked inhibitor of apoptosis protein (XIAP), and provides a summary of the promising approaches currently in development that indicate the future potential of PPI inhibitors in drug discovery.
Figures


References
-
- Bellay J, Michaut M, Kim T, Han S, Colak R, Myers CL, Kim PM. An omics perspective of protein disorder. Mol Biosyst. 2012;8:185–93. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
Miscellaneous

