PrPSc-Specific Antibody Reveals C-Terminal Conformational Differences between Prion Strains
- PMID: 26937029
- PMCID: PMC4859706
- DOI: 10.1128/JVI.00088-16
PrPSc-Specific Antibody Reveals C-Terminal Conformational Differences between Prion Strains
Abstract
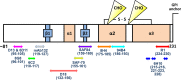
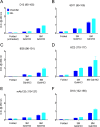
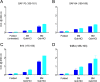
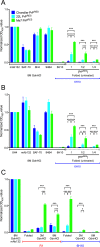
Understanding the structure of PrP(Sc) and its strain variation has been one of the major challenges in prion disease biology. To study the strain-dependent conformations of PrP(Sc), we purified proteinase-resistant PrP(Sc) (PrP(RES)) from mouse brains with three different murine-adapted scrapie strains (Chandler, 22L, and Me7) and systematically tested the accessibility of epitopes of a wide range of anti-PrP and anti-PrP(Sc) specific antibodies by indirect enzyme-linked immunosorbent assay (ELISA). We found that epitopes of most anti-PrP antibodies were hidden in the folded structure of PrP(RES), even though these epitopes are revealed with guanidine denaturation. However, reactivities to a PrP(Sc)-specific conformational C-terminal antibody showed significant differences among the three different prion strains. Our results provide evidence for strain-dependent conformational variation near the C termini of molecules within PrP(Sc) multimers.
Importance: It has long been apparent that prion strains can have different conformations near the N terminus of the PrP(Sc) protease-resistant core. Here, we show that a C-terminal conformational PrP(Sc)-specific antibody reacts differently to three murine-adapted scrapie strains. These results suggest, in turn, that conformational differences in the C terminus of PrP(Sc) also contribute to the phenotypic distinction between prion strains.
Copyright © 2016, American Society for Microbiology. All Rights Reserved.
Figures
Similar articles
-
Novel epitopes identified by anti-PrP monoclonal antibodies produced following immunization of Prnp0/0 Balb/cJ mice with purified scrapie prions.Hybridoma (Larchmt). 2012 Oct;31(5):314-24. doi: 10.1089/hyb.2012.0022. Hybridoma (Larchmt). 2012. PMID: 23098297 Free PMC article.
-
Different antigenicities of the N-terminal region of cellular and scrapie prion proteins.Microbiol Immunol. 2013 Nov;57(11):792-6. doi: 10.1111/1348-0421.12105. Microbiol Immunol. 2013. PMID: 24117858
-
Epitope scanning reveals gain and loss of strain specific antibody binding epitopes associated with the conversion of normal cellular prion to scrapie prion.J Neurochem. 2004 Sep;90(5):1205-17. doi: 10.1111/j.1471-4159.2004.02582.x. J Neurochem. 2004. PMID: 15312175
-
The use of monoclonal antibody epitopes for tagging PrP in conversion experiments.Arch Virol Suppl. 2000;(16):285-90. doi: 10.1007/978-3-7091-6308-5_27. Arch Virol Suppl. 2000. PMID: 11214932 Review.
-
Prion Strain Diversity.Cold Spring Harb Perspect Med. 2016 Dec 1;6(12):a024349. doi: 10.1101/cshperspect.a024349. Cold Spring Harb Perspect Med. 2016. PMID: 27908925 Free PMC article. Review.
Cited by
-
Cofactor and glycosylation preferences for in vitro prion conversion are predominantly determined by strain conformation.PLoS Pathog. 2020 Apr 15;16(4):e1008495. doi: 10.1371/journal.ppat.1008495. eCollection 2020 Apr. PLoS Pathog. 2020. PMID: 32294141 Free PMC article.
-
Challenges and Advances in Antemortem Diagnosis of Human Transmissible Spongiform Encephalopathies.Front Bioeng Biotechnol. 2020 Oct 20;8:585896. doi: 10.3389/fbioe.2020.585896. eCollection 2020. Front Bioeng Biotechnol. 2020. PMID: 33195151 Free PMC article. Review.
-
Enhancement of binding avidity by bivalent binding enables PrPSc-specific detection by anti-PrP monoclonal antibody 132.PLoS One. 2019 Jun 6;14(6):e0217944. doi: 10.1371/journal.pone.0217944. eCollection 2019. PLoS One. 2019. PMID: 31170247 Free PMC article.
-
Prion protein PrP nucleic acid binding and mobilization implicates retroelements as the replicative component of transmissible spongiform encephalopathy.Arch Virol. 2020 Mar;165(3):535-556. doi: 10.1007/s00705-020-04529-2. Epub 2020 Feb 5. Arch Virol. 2020. PMID: 32025859 Free PMC article. Review.
-
Prion-Like Propagation of Post-Translationally Modified Tau in Alzheimer's Disease: A Hypothesis.J Mol Neurosci. 2018 Aug;65(4):480-490. doi: 10.1007/s12031-018-1111-5. Epub 2018 Jul 7. J Mol Neurosci. 2018. PMID: 29982964 Review.
References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

