The RNA Helicase eIF4A Is Required for Sapovirus Translation
- PMID: 26937032
- PMCID: PMC4859703
- DOI: 10.1128/JVI.03174-15
The RNA Helicase eIF4A Is Required for Sapovirus Translation
Abstract
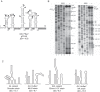
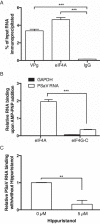
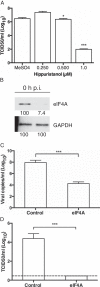
The eukaryotic initiation factor 4A (eIF4A) is a DEAD box helicase that unwinds RNA structure in the 5' untranslated region (UTR) of mRNAs. Here, we investigated the role of eIF4A in porcine sapovirus VPg-dependent translation. Using inhibitors and dominant-negative mutants, we found that eIF4A is required for viral translation and infectivity, suggesting that despite the presence of a very short 5' UTR, eIF4A is required to unwind RNA structure in the sapovirus genome to facilitate virus translation.
Copyright © 2016 Hosmillo et al.
Figures

Similar articles
-
Identification and characterization of hippuristanol-resistant mutants reveals eIF4A1 dependencies within mRNA 5' leader regions.Nucleic Acids Res. 2020 Sep 25;48(17):9521-9537. doi: 10.1093/nar/gkaa662. Nucleic Acids Res. 2020. PMID: 32766783 Free PMC article.
-
eIF4B stimulates eIF4A ATPase and unwinding activities by direct interaction through its 7-repeats region.RNA Biol. 2017 Jan 2;14(1):113-123. doi: 10.1080/15476286.2016.1259782. Epub 2016 Nov 18. RNA Biol. 2017. PMID: 27858515 Free PMC article.
-
Mutational analysis of the DEAD-box RNA helicase eIF4AII characterizes its interaction with transformation suppressor Pdcd4 and eIF4GI.RNA. 2005 Mar;11(3):261-74. doi: 10.1261/rna.7191905. Epub 2005 Jan 20. RNA. 2005. PMID: 15661843 Free PMC article.
-
Eukaryotic initiation factor 4A (eIF4A) during viral infections.Virus Genes. 2019 Jun;55(3):267-273. doi: 10.1007/s11262-019-01641-7. Epub 2019 Feb 22. Virus Genes. 2019. PMID: 30796742 Free PMC article. Review.
-
The DEAD-box helicase eIF4A: paradigm or the odd one out?RNA Biol. 2013 Jan;10(1):19-32. doi: 10.4161/rna.21966. Epub 2012 Sep 20. RNA Biol. 2013. PMID: 22995829 Free PMC article. Review.
Cited by
-
Translational Control during Calicivirus Infection.Viruses. 2016 Apr 20;8(4):104. doi: 10.3390/v8040104. Viruses. 2016. PMID: 27104553 Free PMC article. Review.
-
Long non-coding RNAs in Epstein-Barr virus-related cancer.Cancer Cell Int. 2021 May 25;21(1):278. doi: 10.1186/s12935-021-01986-w. Cancer Cell Int. 2021. PMID: 34034760 Free PMC article. Review.
-
Noroviruses subvert the core stress granule component G3BP1 to promote viral VPg-dependent translation.Elife. 2019 Aug 12;8:e46681. doi: 10.7554/eLife.46681. Elife. 2019. PMID: 31403400 Free PMC article.
-
Caliciviridae Other Than Noroviruses.Viruses. 2019 Mar 21;11(3):286. doi: 10.3390/v11030286. Viruses. 2019. PMID: 30901945 Free PMC article. Review.
-
First complete genome sequences of genogroup V, genotype 3 porcine sapoviruses: common 5'-terminal genomic feature of sapoviruses.Virus Genes. 2017 Dec;53(6):848-855. doi: 10.1007/s11262-017-1481-8. Epub 2017 Jun 22. Virus Genes. 2017. PMID: 28643180
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

