c-MYB is a transcriptional regulator of ESPL1/Separase in BCR-ABL-positive chronic myeloid leukemia
- PMID: 26937281
- PMCID: PMC4774018
- DOI: 10.1186/s40364-016-0059-2
c-MYB is a transcriptional regulator of ESPL1/Separase in BCR-ABL-positive chronic myeloid leukemia
Abstract
Background: Genomic instability and clonal evolution are hallmarks of progressing chronic myeloid leukemia (CML). Recently, we have shown that clonal evolution and blast crisis correlate with altered expression and activity of Separase, a cysteine endopeptidase that is a mitotic key player in chromosomal segregation and centriole duplication. Hyperactivation of Separase in human hematopoietic cells has been linked to a feedback mechanism that posttranslationally stimulates Separase proteolytic activity after imatinib therapy-induced reduction of Separase protein levels.
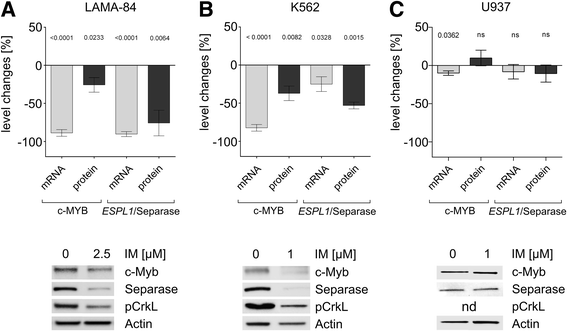
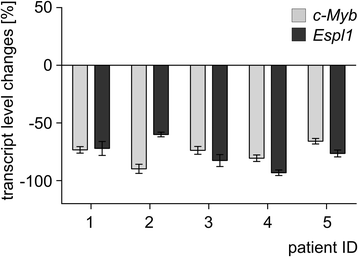
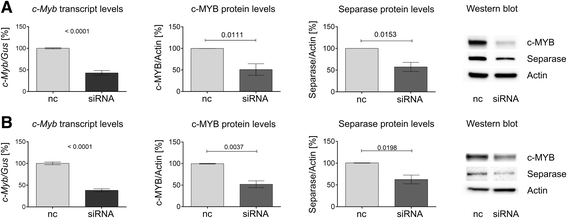
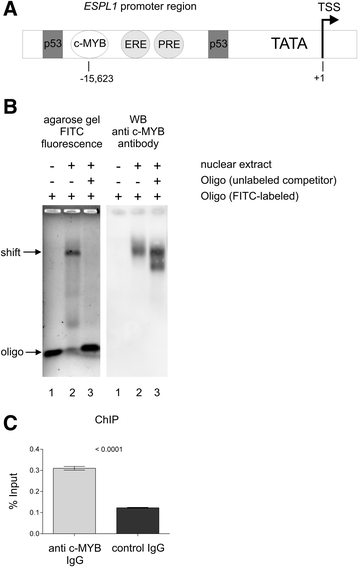
Methods and results: In search for potential therapy-responsive transcriptional mechanisms we have investigated the role of the transcription factor c-MYB for Separase expression in CML cell lines (LAMA-84, K562, BV-173) and in clinical samples. Quantitative RT-PCR and Western blot immunostaining experiments revealed that c-MYB expression levels are decreased in an imatinib-dependent manner and positively correlate with Separase expression levels in cell lines and in clinical CML samples. RNA silencing of c-MYB expression in CML cell lines resulted in reduced Separase protein levels. Gelshift and ChIP assays confirmed that c-MYB binds to a putative c-MYB binding sequence located within the ESPL1 promoter.
Conclusions: Our data suggest that ESPL1/Separase is a regulatory target of c-MYB. Therefore, c-MYB, known to be required for BCR-ABL-dependent transformation of hematopoietic progenitors and leukemogenesis, may also control the Separase-dependent fidelity of mitotic chromosomal segregation and centriole duplication essential for maintenance of genomic stability.
Keywords: BCR-ABL; CML; ESPL1/Separase; Genomic stability; Imatinib; c-MYB.
Figures
References
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
Miscellaneous

