A Heterologous Multiepitope DNA Prime/Recombinant Protein Boost Immunisation Strategy for the Development of an Antiserum against Micrurus corallinus (Coral Snake) Venom
- PMID: 26938217
- PMCID: PMC4777291
- DOI: 10.1371/journal.pntd.0004484
A Heterologous Multiepitope DNA Prime/Recombinant Protein Boost Immunisation Strategy for the Development of an Antiserum against Micrurus corallinus (Coral Snake) Venom
Abstract
Background: Envenoming by coral snakes (Elapidae: Micrurus), although not abundant, represent a serious health threat in the Americas, especially because antivenoms are scarce. The development of adequate amounts of antielapidic serum for the treatment of accidents caused by snakes like Micrurus corallinus is a challenging task due to characteristics such as low venom yield, fossorial habit, relatively small sizes and ophiophagous diet. These features make it difficult to capture and keep these snakes in captivity for venom collection. Furthermore, there are reports of antivenom scarcity in USA, leading to an increase in morbidity and mortality, with patients needing to be intubated and ventilated while the toxin wears off. The development of an alternative method for the production of an antielapidic serum, with no need for snake collection and maintenance in captivity, would be a plausible solution for the antielapidic serum shortage.
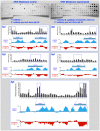
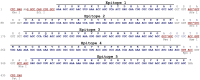
Methods and findings: In this work we describe the mapping, by the SPOT-synthesis technique, of potential B-cell epitopes from five putative toxins from M. corallinus, which were used to design two multiepitope DNA strings for the genetic immunisation of female BALB/c mice. Results demonstrate that sera obtained from animals that were genetically immunised with these multiepitope constructs, followed by booster doses of recombinant proteins lead to a 60% survival in a lethal dose neutralisation assay.
Conclusion: Here we describe that the genetic immunisation with a synthetic multiepitope gene followed by booster doses with recombinant protein is a promising approach to develop an alternative antielapidic serum against M. corallinus venom without the need of collection and the very challenging maintenance of these snakes in captivity.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures






References
-
- Warrell DA. Epidemiology, clinical features and management of snake bites in Central and South America Venomous Reptiles of the Western Hemisphere: Cornell University Press; 2004. p. 709–61.
-
- Bucaretchi F, Hyslop S, Vieira RJ, Toledo AS, Madureira PR, de Capitani EM. Bites by coral snakes (Micrurus spp.) in Campinas, State of Sao Paulo, Southeastern Brazil. Revista do Instituto de Medicina Tropical de Sao Paulo. 2006;48(3):141–5. - PubMed
-
- Roze JA. Coral snakes of the Americas—biology, identification, and venoms: Krieger Pub Co; Original ed edition; 1996. 340 p.
-
- Cardoso JLC, França FOdS, Wen FH, Malaque CMSA, V H Jr.. Animais peçonhentos no Brasil: biologia clínica e terapêutica dos acidentes. 2 ed: Sarvier; 2009.
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

