Molecular Characterization of the Cercosporin Biosynthetic Pathway in the Fungal Plant Pathogen Cercospora nicotianae
- PMID: 26938470
- PMCID: PMC5129747
- DOI: 10.1021/jacs.6b00633
Molecular Characterization of the Cercosporin Biosynthetic Pathway in the Fungal Plant Pathogen Cercospora nicotianae
Abstract
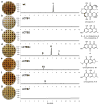
Perylenequinones are a class of photoactivated polyketide mycotoxins produced by fungal plant pathogens that notably produce reactive oxygen species with visible light. The best-studied perylenequinone is cercosporin-a product of the Cercospora species. While the cercosporin biosynthetic gene cluster has been described in the tobacco pathogen Cercospora nicotianae, little is known of the metabolite's biosynthesis. Furthermore, in vitro investigations of the polyketide synthase central to cercosporin biosynthesis identified the naphthopyrone nor-toralactone as its direct product-an observation in conflict with published biosynthetic proposals. Here, we present an alternative biosynthetic pathway to cercosporin based on metabolites characterized from a series of biosynthetic gene knockouts. We show that nor-toralactone is the key polyketide intermediate and the substrate for the unusual didomain protein CTB3. We demonstrate the unique oxidative cleavage activity of the CTB3 monooxygenase domain in vitro. These data advance our understanding of perylenequinone biosynthesis and expand the biochemical repertoire of flavin-dependent monooxygenases.
Figures




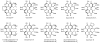
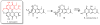

References
-
- Daub ME. Phytopathology. 1981;71:213–213.
-
- Daub ME, Herrero S, Chung KR. FEMS Microbiol Lett. 2005;252:197–206. - PubMed
-
- Kuyama S, Tamura T. J Am Chem Soc. 1957;79:5725–5726.
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

