Identifying Likely Transmission Pathways within a 10-Year Community Outbreak of Tuberculosis by High-Depth Whole Genome Sequencing
- PMID: 26938641
- PMCID: PMC4777479
- DOI: 10.1371/journal.pone.0150550
Identifying Likely Transmission Pathways within a 10-Year Community Outbreak of Tuberculosis by High-Depth Whole Genome Sequencing
Abstract
Background: Improved tuberculosis control and the need to contain the spread of drug-resistant strains provide a strong rationale for exploring tuberculosis transmission dynamics at the population level. Whole-genome sequencing provides optimal strain resolution, facilitating detailed mapping of potential transmission pathways.
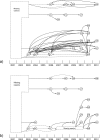
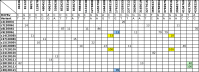
Methods: We sequenced 22 isolates from a Mycobacterium tuberculosis cluster in New South Wales, Australia, identified during routine 24-locus mycobacterial interspersed repetitive unit typing. Following high-depth paired-end sequencing using the Illumina HiSeq 2000 platform, two independent pipelines were employed for analysis, both employing read mapping onto reference genomes as well as de novo assembly, to control biases in variant detection. In addition to single-nucleotide polymorphisms, the analyses also sought to identify insertions, deletions and structural variants.
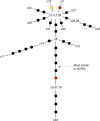
Results: Isolates were highly similar, with a distance of 13 variants between the most distant members of the cluster. The most sensitive analysis classified the 22 isolates into 18 groups. Four of the isolates did not appear to share a recent common ancestor with the largest clade; another four isolates had an uncertain ancestral relationship with the largest clade.
Conclusion: Whole genome sequencing, with analysis of single-nucleotide polymorphisms, insertions, deletions, structural variants and subpopulations, enabled the highest possible level of discrimination between cluster members, clarifying likely transmission pathways and exposing the complexity of strain origin. The analysis provides a basis for targeted public health intervention and enhanced classification of future isolates linked to the cluster.
Conflict of interest statement
Figures


References
-
- World Health Organization. Global tuberculosis report 2014 [Internet]. Geneva, Switzerland: World Health Organization; 2014. Available: http://www.who.int/tb/publications/global_report/en/.
MeSH terms
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical

