Identification of mRNA isoform switching in breast cancer
- PMID: 26939613
- PMCID: PMC4778320
- DOI: 10.1186/s12864-016-2521-9
Identification of mRNA isoform switching in breast cancer
Abstract
Background: Alternative splicing provides a major mechanism to generate protein diversity. Increasing evidence suggests a link of dysregulation of splicing associated with cancer. While previous genomic-based studies demonstrated the expression of a handful of tumor-specific isoforms, genome-wide alterations in the balance between isoforms and cancer subtypes is understudied.
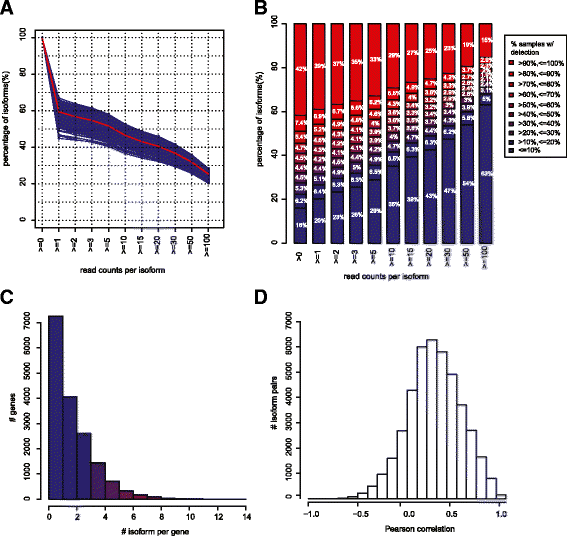
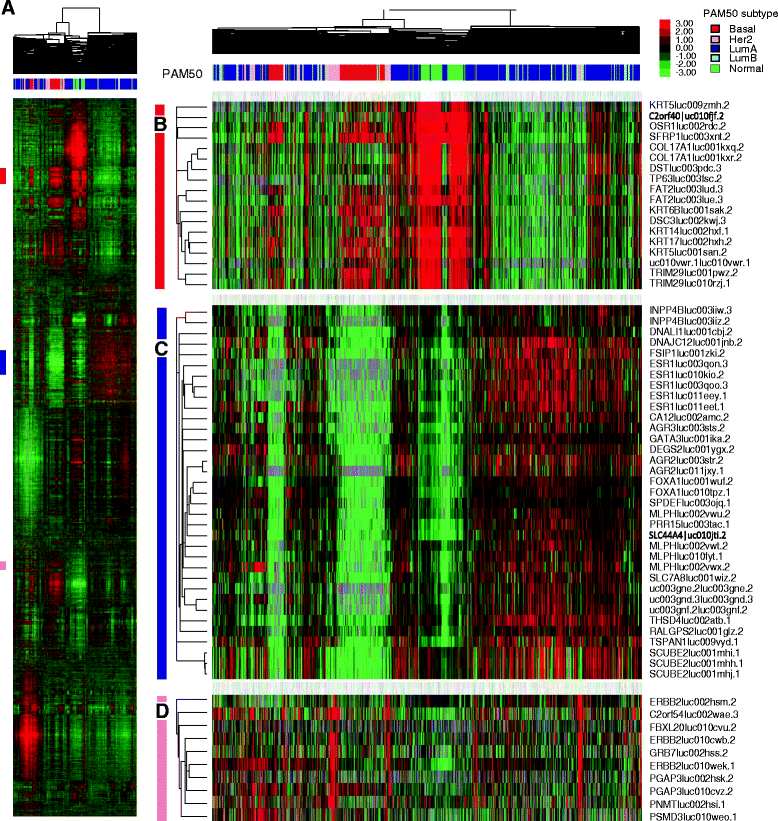
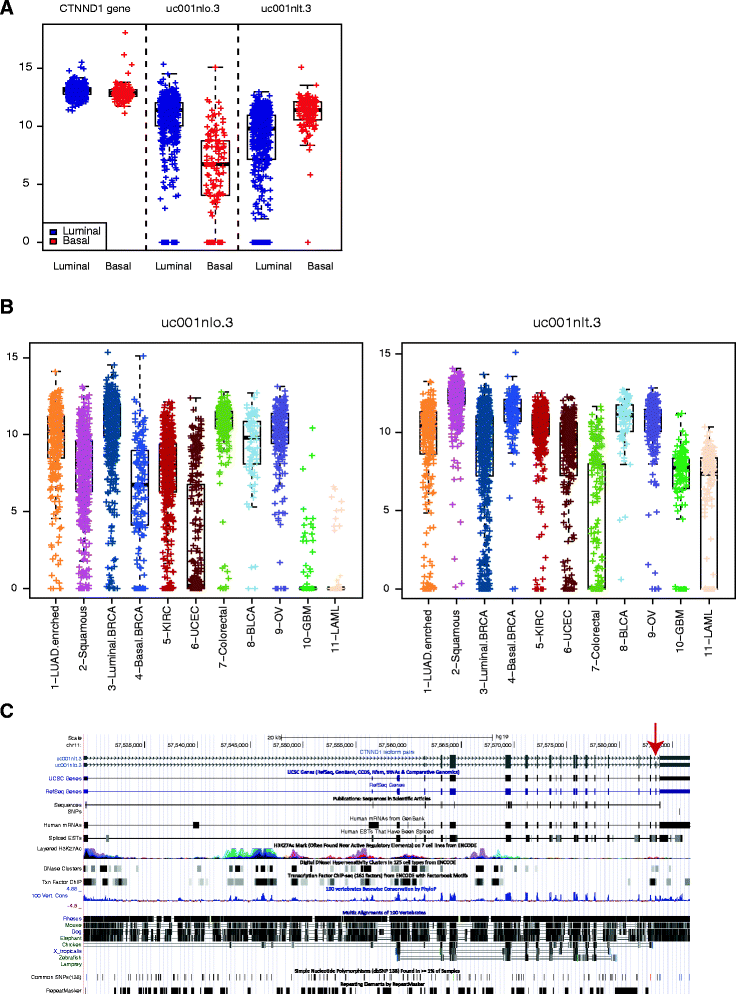
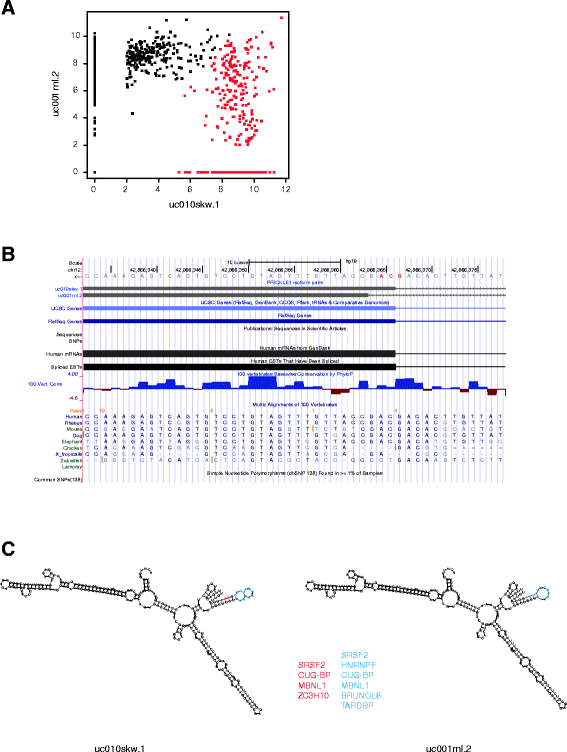
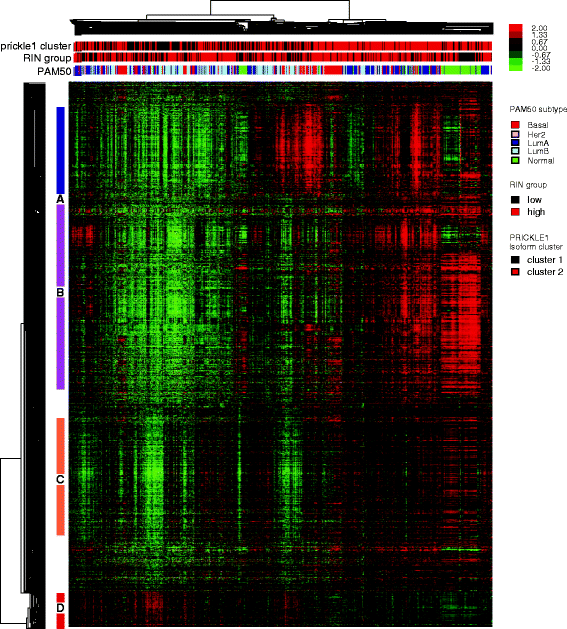
Result: We systematically analyzed the isoform-level expression patterns and isoform switching events of 819 breast tumor and normal samples assayed by mRNA-seq from TCGA project. On average, 2.2 isoforms per gene were detected and 67.5 % of detected genes (i.e. expressed) showed 1-2 isoforms only. While the majority of isoforms for a given gene were positively correlated with each other and the overall gene level, 470 pairs of isoforms displayed an inverse correlation suggesting a switching event. Most of the isoform switching events were associated with molecular subtypes, including a Basal-like-associated switching in CTNND1. 88 genes showed switching independent of subtypes, among which the isoform pattern of PRICKLE1 was associated with a large genomic signature of biological significance.
Conclusion: Our results reveal that the majority of genes do not undergo complex mRNA splicing within breast cancers, and that there is a general concordance in isoform and gene expression levels in breast tumors. We identified hundreds of isoform switching events across breast tumors, most of which were associated with differences in tumor subtypes. As exemplified by the detailed analysis of CTNND1 and PRICKLE1, these isoform switching events potentially provide new insights into the post-transcriptional regulatory mechanisms of tumor subtypes and cancer biology.
Figures
References
-
- Oltean S, Bates DO. Hallmarks of alternative splicing in cancer. Oncogene. 2014;33(46):5311–8. - PubMed
-
- Venables JP, Klinck R, Bramard A, Inkel L, Dufresne-Martin G, Koh C, Gervais-Bird J, Lapointe E, Froehlich U, Durand M, Gendron D, Brosseau J-P, Thibault P, Lucier J-F, Tremblay K, Prinos P, Wellinger RJ, Chabot B, Rancourt C, Elela SA. Identification of alternative splicing markers for breast cancer. Cancer Res. 2008;68:9525–31. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Research Materials
Miscellaneous

