Genetic alterations of triple negative breast cancer by targeted next-generation sequencing and correlation with tumor morphology
- PMID: 26939876
- PMCID: PMC4848211
- DOI: 10.1038/modpathol.2016.39
Genetic alterations of triple negative breast cancer by targeted next-generation sequencing and correlation with tumor morphology
Abstract
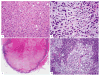
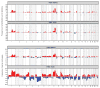
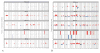
Triple negative breast cancer represents a heterogeneous group of breast carcinomas, both at the histologic and genetic level. Although recent molecular studies have comprehensively characterized the genetic landscape of these tumors, few have integrated a detailed histologic examination into the analysis. In this study, we defined the genetic alterations in 39 triple negative breast cancers using a high-depth targeted massively parallel sequencing assay and correlated the findings with a detailed morphologic analysis. We obtained representative frozen tissue of primary triple negative breast cancers from patients treated at our institution between 2002 and 2010. We characterized tumors according to their histologic subtype and morphologic features. DNA was extracted from paired frozen primary tumor and normal tissue samples and was subjected to a targeted massively parallel sequencing platform comprising 229 cancer-associated genes common across all experiments. The average number of non-synonymous mutations was 3 (range 0-10) per case. The most frequent somatic alterations were mutations in TP53 (74%) and PIK3CA (10%) and MYC amplifications (26%). Triple negative breast cancers with apocrine differentiation less frequently harbored TP53 mutations (25%) and MYC gains (0%), and displayed a high mutation frequency in PIK3CA and other PI3K signaling pathway-related genes (75%). Using a targeted massively parallel sequencing platform, we identified the key somatic genetic alterations previously reported in triple negative breast cancers. Furthermore, our findings show that triple negative breast cancers with apocrine differentiation constitute a distinct subset, characterized by a high frequency of PI3K pathway alterations similar to luminal subtypes of breast cancer.
Conflict of interest statement
Conflict of interest
The authors declare no conflict of interest.
Figures


References
-
- Dent R, Trudeau M, Pritchard KI, et al. Triple-negative breast cancer: clinical features and patterns of recurrence. Clin Cancer Res. 2007;13:4429–4434. - PubMed
-
- Livasy CA, Karaca G, Nanda R, et al. Phenotypic evaluation of the basal-like subtype of invasive breast carcinoma. Mod Pathol. 2006;19:264–271. - PubMed
-
- Thike AA, Cheok PY, Jara-Lazaro AR, Tan B, Tan P, Tan PH. Triple-negative breast cancer: clinicopathological characteristics and relationship with basal-like breast cancer. Mod Pathol. 2010;23:123–133. - PubMed
-
- Weigelt B, Kreike B, Reis-Filho JS. Metaplastic breast carcinomas are basal-like breast cancers: a genomic profiling analysis. Breast Cancer Res Treat. 2009;117:273–280. - PubMed
-
- Geyer FC, Weigelt B, Natrajan R, et al. Molecular analysis reveals a genetic basis for the phenotypic diversity of metaplastic breast carcinomas. J Pathol. 2010;220:562–573. - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials
Miscellaneous

