The orientation of transcription factor binding site motifs in gene promoter regions: does it matter?
- PMID: 26939991
- PMCID: PMC4778318
- DOI: 10.1186/s12864-016-2549-x
The orientation of transcription factor binding site motifs in gene promoter regions: does it matter?
Erratum in
-
Erratum to: 'The orientation of transcription factor binding site motifs in gene promoter regions: does it matter?'.BMC Genomics. 2016 Apr 28;17:310. doi: 10.1186/s12864-016-2630-5. BMC Genomics. 2016. PMID: 27126049 Free PMC article. No abstract available.
Abstract
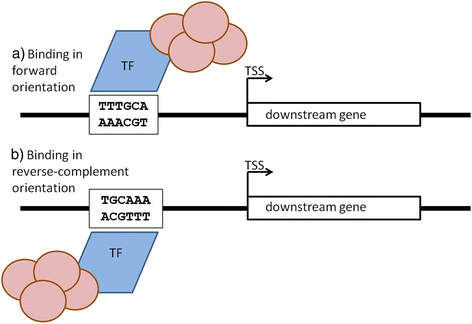
Background: Gene expression is to large degree regulated by the specific binding of protein transcription factors to cis-regulatory transcription factor binding sites in gene promoter regions. Despite the identification of hundreds of binding site sequence motifs, the question as to whether motif orientation matters with regard to the gene expression regulation of the respective downstream genes appears surprisingly underinvestigated.
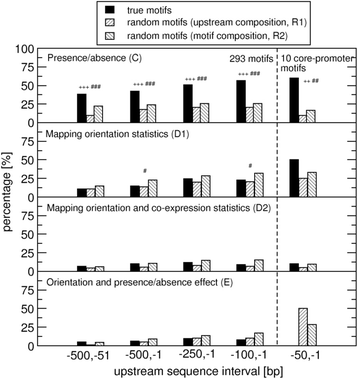
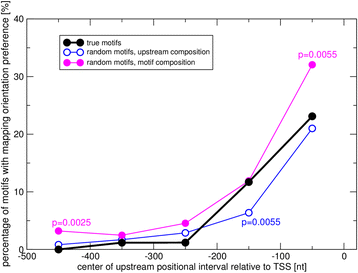
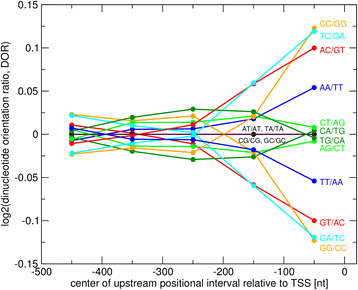
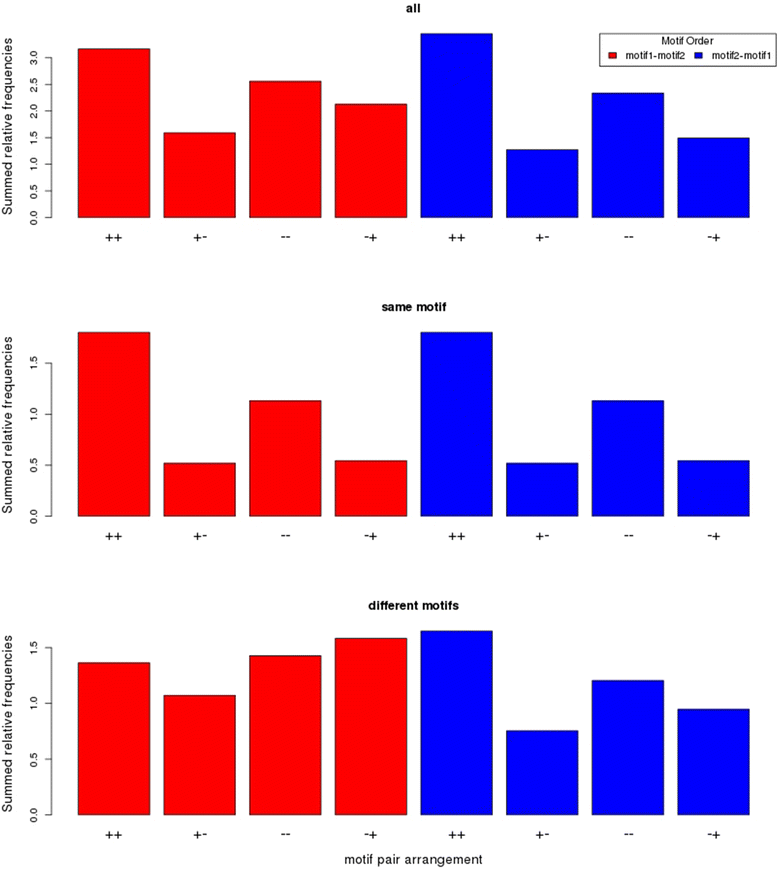
Results: We pursued a statistical approach by probing 293 reported non-palindromic transcription factor binding site and ten core promoter motifs in Arabidopsis thaliana for evidence of any relevance of motif orientation based on mapping statistics and effects on the co-regulation of gene expression of the respective downstream genes. Although positional intervals closer to the transcription start site (TSS) were found with increased frequencies of motifs exhibiting orientation preference, a corresponding effect with regard to gene expression regulation as evidenced by increased co-expression of genes harboring the favored orientation in their upstream sequence could not be established. Furthermore, we identified an intrinsic orientational asymmetry of sequence regions close to the TSS as the likely source of the identified motif orientation preferences. By contrast, motif presence irrespective of orientation was found associated with pronounced effects on gene expression co-regulation validating the pursued approach. Inspecting motif pairs revealed statistically preferred orientational arrangements, but no consistent effect with regard to arrangement-dependent gene expression regulation was evident.
Conclusions: Our results suggest that for the motifs considered here, either no specific orientation rendering them functional across all their instances exists with orientational requirements instead depending on gene-locus specific additional factors, or that the binding orientation of transcription factors may generally not be relevant, but rather the event of binding itself.
Figures
References
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

