Computational methods for ubiquitination site prediction using physicochemical properties of protein sequences
- PMID: 26940649
- PMCID: PMC4778322
- DOI: 10.1186/s12859-016-0959-z
Computational methods for ubiquitination site prediction using physicochemical properties of protein sequences
Abstract
Background: Ubiquitination is a very important process in protein post-translational modification, which has been widely investigated by biology scientists and researchers. Different experimental and computational methods have been developed to identify the ubiquitination sites in protein sequences. This paper aims at exploring computational machine learning methods for the prediction of ubiquitination sites using the physicochemical properties (PCPs) of amino acids in the protein sequences.
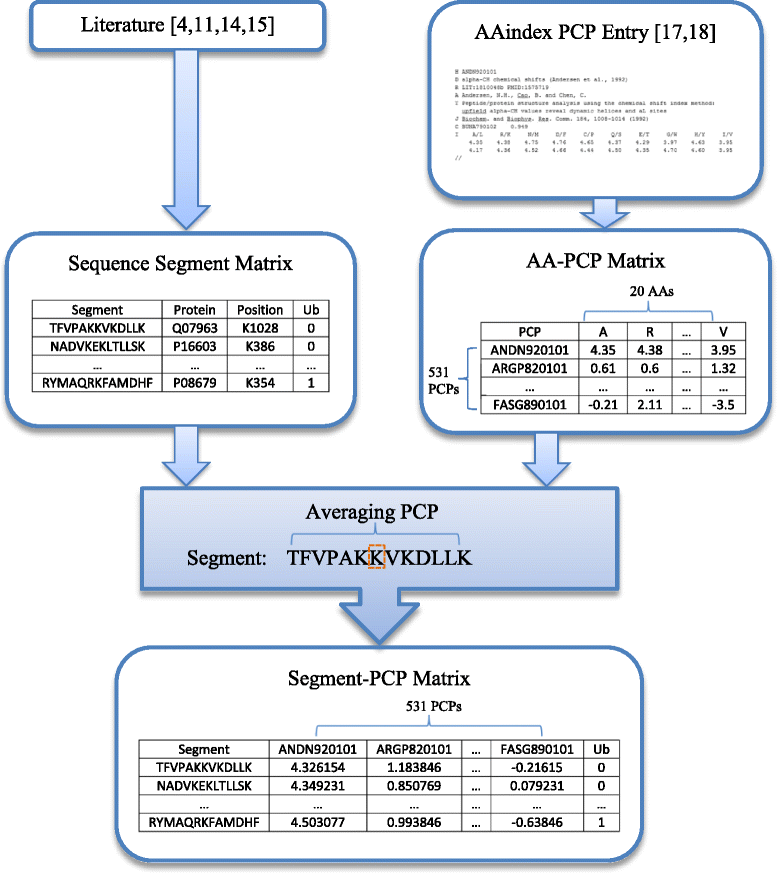
Results: We first establish six different ubiquitination data sets, whose records contain both ubiquitination sites and non-ubiquitination sites in variant numbers of protein sequence segments. In particular, to establish such data sets, protein sequence segments are extracted from the original protein sequences used in four published papers on ubiquitination, while 531 PCP features of each extracted protein sequence segment are calculated based on PCP values from AAindex (Amino Acid index database) by averaging PCP values of all amino acids on each segment. Various computational machine-learning methods, including four Bayesian network methods (i.e., Naïve Bayes (NB), Feature Selection NB (FSNB), Model Averaged NB (MANB), and Efficient Bayesian Multivariate Classifier (EBMC)) and three regression methods (i.e., Support Vector Machine (SVM), Logistic Regression (LR), and Least Absolute Shrinkage and Selection Operator (LASSO)), are then applied to the six established segment-PCP data sets. Five-fold cross-validation and the Area Under Receiver Operating Characteristic Curve (AUROC) are employed to evaluate the ubiquitination prediction performance of each method. Results demonstrate that the PCP data of protein sequences contain information that could be mined by machine learning methods for ubiquitination site prediction. The comparative results show that EBMC, SVM and LR perform better than other methods, and EBMC is the only method that can get AUCs greater than or equal to 0.6 for the six established data sets. Results also show EBMC tends to perform better for larger data.
Conclusions: Machine learning methods have been employed for the ubiquitination site prediction based on physicochemical properties of amino acids on protein sequences. Results demonstrate the effectiveness of using machine learning methodology to mine information from PCP data concerning protein sequences, as well as the superiority of EBMC, SVM and LR (especially EBMC) for the ubiquitination prediction compared to other methods.
Figures
References
-
- The 2004 Nobel Prize in Chemistry - Popular Information. Nobelprize.org. Nobel Media AB 2014. Available online at www.nobelprize.org/nobel_prizes/chemistry/laureates/2004/popular.html. Accessed 26 Nov 2014.
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous