Activation of proto-oncogenes by disruption of chromosome neighborhoods
- PMID: 26940867
- PMCID: PMC4884612
- DOI: 10.1126/science.aad9024
Activation of proto-oncogenes by disruption of chromosome neighborhoods
Abstract
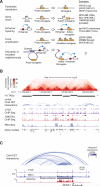
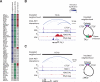
Oncogenes are activated through well-known chromosomal alterations such as gene fusion, translocation, and focal amplification. In light of recent evidence that the control of key genes depends on chromosome structures called insulated neighborhoods, we investigated whether proto-oncogenes occur within these structures and whether oncogene activation can occur via disruption of insulated neighborhood boundaries in cancer cells. We mapped insulated neighborhoods in T cell acute lymphoblastic leukemia (T-ALL) and found that tumor cell genomes contain recurrent microdeletions that eliminate the boundary sites of insulated neighborhoods containing prominent T-ALL proto-oncogenes. Perturbation of such boundaries in nonmalignant cells was sufficient to activate proto-oncogenes. Mutations affecting chromosome neighborhood boundaries were found in many types of cancer. Thus, oncogene activation can occur via genetic alterations that disrupt insulated neighborhoods in malignant cells.
Copyright © 2016, American Association for the Advancement of Science.
Figures


Comment in
-
CANCER. The oncogene makes its escape.Science. 2016 Mar 25;351(6280):1398-9. doi: 10.1126/science.aaf5542. Science. 2016. PMID: 27013717 Free PMC article. No abstract available.
References
-
- Vogelstein B, Kinzler KW. Cancer genes and the pathways they control. Nature medicine. 2004 Aug;10:789. - PubMed
-
- Garraway LA, Lander ES. Lessons from the cancer genome. Cell. 2013 Mar 28;153:17. - PubMed
-
- Croce CM. Oncogenes and cancer. The New England journal of medicine. 2008 Jan 31;358:502. - PubMed
Publication types
MeSH terms
Associated data
- Actions
Grants and funding
- U01 R01 AI 117839/AI/NIAID NIH HHS/United States
- T32 GM007287/GM/NIGMS NIH HHS/United States
- R01 MH104610/MH/NIMH NIH HHS/United States
- AI120766/AI/NIAID NIH HHS/United States
- P01 CA109901/CA/NCI NIH HHS/United States
- U01 DA 040588/DA/NIDA NIH HHS/United States
- R01 HG003143/HG/NHGRI NIH HHS/United States
- R01 GM 112720/GM/NIGMS NIH HHS/United States
- U54 DK107980/DK/NIDDK NIH HHS/United States
- P30 CA014051/CA/NCI NIH HHS/United States
- R01 AI120766/AI/NIAID NIH HHS/United States
- R25 HG007631/HG/NHGRI NIH HHS/United States
- R01 NS088538/NS/NINDS NIH HHS/United States
- R37 HD045022/HD/NICHD NIH HHS/United States
- T32 GM087237/GM/NIGMS NIH HHS/United States
- NS088538/NS/NINDS NIH HHS/United States
- U01 HG007910/HG/NHGRI NIH HHS/United States
- Howard Hughes Medical Institute/United States
- R01 HG002668/HG/NHGRI NIH HHS/United States
- U54 HG007010/HG/NHGRI NIH HHS/United States
- HG002668/HG/NHGRI NIH HHS/United States
- MH104610/MH/NIMH NIH HHS/United States
- U54 CA193419/CA/NCI NIH HHS/United States
- CA109901/CA/NCI NIH HHS/United States
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

