Defects in Peroxisomal 6-Phosphogluconate Dehydrogenase Isoform PGD2 Prevent Gametophytic Interaction in Arabidopsis thaliana
- PMID: 26941195
- PMCID: PMC4854672
- DOI: 10.1104/pp.15.01301
Defects in Peroxisomal 6-Phosphogluconate Dehydrogenase Isoform PGD2 Prevent Gametophytic Interaction in Arabidopsis thaliana
Abstract
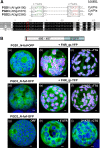
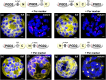
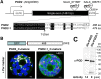
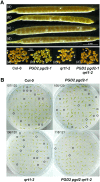
We studied the localization of 6-phosphogluconate dehydrogenase (PGD) isoforms of Arabidopsis (Arabidopsis thaliana). Similar polypeptide lengths of PGD1, PGD2, and PGD3 obscured which isoform may represent the cytosolic and/or plastidic enzyme plus whether PGD2 with a peroxisomal targeting motif also might target plastids. Reporter-fusion analyses in protoplasts revealed that, with a free N terminus, PGD1 and PGD3 accumulate in the cytosol and chloroplasts, whereas PGD2 remains in the cytosol. Mutagenesis of a conserved second ATG enhanced the plastidic localization of PGD1 and PGD3 but not PGD2. Amino-terminal deletions of PGD2 fusions with a free C terminus resulted in peroxisomal import after dimerization, and PGD2 could be immunodetected in purified peroxisomes. Repeated selfing of pgd2 transfer (T-)DNA alleles yielded no homozygous mutants, although siliques and seeds of heterozygous plants developed normally. Detailed analyses of the C-terminally truncated PGD2-1 protein showed that peroxisomal import and catalytic activity are abolished. Reciprocal backcrosses of pgd2-1 suggested that missing PGD activity in peroxisomes primarily affects the male gametophyte. Tetrad analyses in the quartet1-2 background revealed that pgd2-1 pollen is vital and in vitro germination normal, but pollen tube growth inside stylar tissues appeared less directed. Mutual gametophytic sterility was overcome by complementation with a genomic construct but not with a version lacking the first ATG. These analyses showed that peroxisomal PGD2 activity is required for guided growth of the male gametophytes and pollen tube-ovule interaction. Our report finally demonstrates an essential role of oxidative pentose-phosphate pathway reactions in peroxisomes, likely needed to sustain critical levels of nitric oxide and/or jasmonic acid, whose biosynthesis both depend on NADPH provision.
© 2016 American Society of Plant Biologists. All Rights Reserved.
Figures





References
-
- Alexander MP. (1969) Differential staining of aborted and nonaborted pollen. Stain Technol 44: 117–122 - PubMed
-
- Alonso JM, Stepanova AN, Leisse TJ, Kim CJ, Chen H, Shinn P, Stevenson DK, Zimmerman J, Barajas P, Cheuk R, et al. (2003) Genome-wide insertional mutagenesis of Arabidopsis thaliana. Science 301: 653–657 - PubMed
-
- Barroso JB, Corpas FJ, Carreras A, Sandalio LM, Valderrama R, Palma JM, Lupiáñez JA, del Río LA (1999) Localization of nitric-oxide synthase in plant peroxisomes. J Biol Chem 274: 36729–36733 - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous

