Cryo-EM structure of a native, fully glycosylated, cleaved HIV-1 envelope trimer
- PMID: 26941313
- PMCID: PMC5001164
- DOI: 10.1126/science.aad2450
Cryo-EM structure of a native, fully glycosylated, cleaved HIV-1 envelope trimer
Abstract
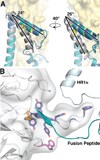
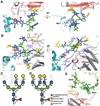
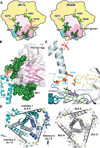
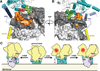
The envelope glycoprotein trimer (Env) on the surface of HIV-1 recognizes CD4(+) T cells and mediates viral entry. During this process, Env undergoes substantial conformational rearrangements, making it difficult to study in its native state. Soluble stabilized trimers have provided valuable insights into the Env structure, but they lack the hydrophobic membrane proximal external region (MPER, an important target of broadly neutralizing antibodies), the transmembrane domain, and the cytoplasmic tail. Here we present (i) a cryogenic electron microscopy (cryo-EM) structure of a clade B virus Env, which lacks only the cytoplasmic tail and is stabilized by the broadly neutralizing antibody PGT151, at a resolution of 4.2 angstroms and (ii) a reconstruction of this form of Env in complex with PGT151 and MPER-targeting antibody 10E8 at a resolution of 8.8 angstroms. These structures provide new insights into the wild-type Env structure.
Copyright © 2016, American Association for the Advancement of Science.
Figures


References
-
- Julien JP, Lee JH, Cupo A, Murin CD, Derking R, Hoffenberg S, Caulfield MJ, King CR, Marozsan AJ, Klasse PJ, Sanders RW, Moore JP, Wilson IA, Ward AB. Asymmetric recognition of the HIV-1 trimer by broadly neutralizing antibody PG9. Proceedings of the National Academy of Sciences of the United States of America. 2013;110:4351–4356. published online EpubMar 12. - PMC - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials

