Regulatory evolution of innate immunity through co-option of endogenous retroviruses
- PMID: 26941318
- PMCID: PMC4887275
- DOI: 10.1126/science.aad5497
Regulatory evolution of innate immunity through co-option of endogenous retroviruses
Abstract
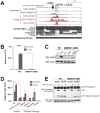
Endogenous retroviruses (ERVs) are abundant in mammalian genomes and contain sequences modulating transcription. The impact of ERV propagation on the evolution of gene regulation remains poorly understood. We found that ERVs have shaped the evolution of a transcriptional network underlying the interferon (IFN) response, a major branch of innate immunity, and that lineage-specific ERVs have dispersed numerous IFN-inducible enhancers independently in diverse mammalian genomes. CRISPR-Cas9 deletion of a subset of these ERV elements in the human genome impaired expression of adjacent IFN-induced genes and revealed their involvement in the regulation of essential immune functions, including activation of the AIM2 inflammasome. Although these regulatory sequences likely arose in ancient viruses, they now constitute a dynamic reservoir of IFN-inducible enhancers fueling genetic innovation in mammalian immune defenses.
Copyright © 2016, American Association for the Advancement of Science.
Figures

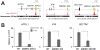

Comment in
-
GENETICS. A copy-and-paste gene regulatory network.Science. 2016 Mar 4;351(6277):1029-30. doi: 10.1126/science.aaf2977. Science. 2016. PMID: 26941305 No abstract available.
-
Evolution: Endogenous Viruses Provide Shortcuts in Antiviral Immunity.Curr Biol. 2016 May 23;26(10):R427-9. doi: 10.1016/j.cub.2016.03.072. Curr Biol. 2016. PMID: 27218853
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

