Multiplexed protein-DNA cross-linking: Scrunching in transcription start site selection
- PMID: 26941320
- PMCID: PMC4797950
- DOI: 10.1126/science.aad6881
Multiplexed protein-DNA cross-linking: Scrunching in transcription start site selection
Abstract
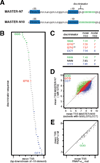
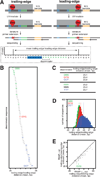
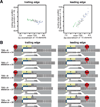
In bacterial transcription initiation, RNA polymerase (RNAP) selects a transcription start site (TSS) at variable distances downstream of core promoter elements. Using next-generation sequencing and unnatural amino acid-mediated protein-DNA cross-linking, we have determined, for a library of 4(10) promoter sequences, the TSS, the RNAP leading-edge position, and the RNAP trailing-edge position. We find that a promoter element upstream of the TSS, the "discriminator," participates in TSS selection, and that, as the TSS changes, the RNAP leading-edge position changes, but the RNAP trailing-edge position does not change. Changes in the RNAP leading-edge position, but not the RNAP trailing-edge position, are a defining hallmark of the "DNA scrunching" that occurs concurrent with RNA synthesis in initial transcription. We propose that TSS selection involves DNA scrunching prior to RNA synthesis.
Copyright © 2016, American Association for the Advancement of Science.
Figures

References
Publication types
MeSH terms
Substances
Grants and funding
- R01 GM041376/GM/NIGMS NIH HHS/United States
- T32 GM007215/GM/NIGMS NIH HHS/United States
- R37 GM041376/GM/NIGMS NIH HHS/United States
- R01 GM037048/GM/NIGMS NIH HHS/United States
- R01 GM115910/GM/NIGMS NIH HHS/United States
- P30 EB009998/EB/NIBIB NIH HHS/United States
- GM37048/GM/NIGMS NIH HHS/United States
- P41 GM111244/GM/NIGMS NIH HHS/United States
- R37 GM037048/GM/NIGMS NIH HHS/United States
- GM115910/GM/NIGMS NIH HHS/United States
- R01 GM088343/GM/NIGMS NIH HHS/United States
- GM041376/GM/NIGMS NIH HHS/United States
- GM088343/GM/NIGMS NIH HHS/United States
LinkOut - more resources
Full Text Sources
Other Literature Sources

