bla CTX-M-152, a Novel Variant of CTX-M-group-25, Identified in a Study Performed on the Prevalence of Multidrug Resistance among Natural Inhabitants of River Yamuna, India
- PMID: 26941715
- PMCID: PMC4762991
- DOI: 10.3389/fmicb.2016.00176
bla CTX-M-152, a Novel Variant of CTX-M-group-25, Identified in a Study Performed on the Prevalence of Multidrug Resistance among Natural Inhabitants of River Yamuna, India
Abstract
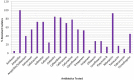
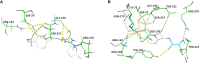
Natural environment influenced by anthropogenic activities creates selective pressure for acquisition and spread of resistance genes. In this study, we determined the prevalence of Extended Spectrum β-Lactamases producing gram negative bacteria from the River Yamuna, India, and report the identification and characterization of a novel CTX-M gene variant bla CTX-M-152 . Of the total 230 non-duplicate isolates obtained from collected water samples, 40 isolates were found positive for ESBL production through Inhibitor-Potentiation Disc Diffusion test. Based on their resistance profile, 3% were found exhibiting pandrug resistance (PDR), 47% extensively drug resistance (XDR), and remaining 50% showing multidrug resistant (MDR). Following screening and antimicrobial profiling, characterization of ESBLs (bla TEM and bla CTX-M ), and mercury tolerance determinants (merP, merT, and merB) were performed. In addition to abundance of bla TEM-116 (57.5%) and bla CTX-M-15 (37.5%), bacteria were also found to harbor other variants of ESBLs like bla CTX-M-71 (5%), bla CTX-M-3 (7.5%), bla CTX-M-32 (2.5%), bla CTX-M-152 (7.5%), bla CTX-M-55 (2.5%), along with some non-ESBLs; bla TEM-1 (25%) and bla OXY (5%). Additionally, co-occurrence of mercury tolerance genes were observed among 40% of isolates. In silico studies of the new variant, bla CTX-M-152 were conducted through modeling for the generation of structure followed by docking to determine its catalytic profile. CTX-M-152 was found to be an out-member of CTX-M-group-25 due to Q26H, T154A, G89D, P99S, and D146G substitutions. Five residues Ser70, Asn132, Ser237, Gly238, and Arg273 were found responsible for positioning of cefotaxime into the active site through seven H-bonds with binding energy of -7.6 Kcal/mol. Despite small active site, co-operative interactions of Ser237 and Arg276 were found actively contributing to its high catalytic efficiency. To the best of our knowledge, this is the first report of bla CTX-M-152 of CTX-M-group-25 from Indian subcontinent. Taking a note of bacteria harboring such high proportion of multidrug and mercury resistance determinants, their presence in natural water resources employed for human consumption increases the chances of potential risk to human health. Hence, deeper insights into mechanisms pertaining to resistance development are required to frame out strategies to tackle the situation and prevent acquisition and dissemination of resistance determinants so as to combat the escalating burden of infectious diseases.
Keywords: ESBL; antibiotics; mechanisms of resistance; polluted environment; resistance genes.
Figures

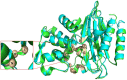

Similar articles
-
Occurrence of OXA-48 Carbapenemase and Other β-Lactamase Genes in ESBL-Producing Multidrug Resistant Escherichia coli from Dogs and Cats in the United States, 2009-2013.Front Microbiol. 2016 Jul 11;7:1057. doi: 10.3389/fmicb.2016.01057. eCollection 2016. Front Microbiol. 2016. PMID: 27462301 Free PMC article.
-
Aeromonas spp. simultaneously harbouring bla(CTX-M-15), bla(SHV-12), bla(PER-1) and bla(FOX-2), in wild-growing Mediterranean mussel (Mytilus galloprovincialis) from Adriatic Sea, Croatia.Int J Food Microbiol. 2013 Sep 2;166(2):301-8. doi: 10.1016/j.ijfoodmicro.2013.07.010. Epub 2013 Jul 18. Int J Food Microbiol. 2013. PMID: 23973842
-
High Prevalence of Drug Resistance and Class 1 Integrons in Escherichia coli Isolated From River Yamuna, India: A Serious Public Health Risk.Front Microbiol. 2021 Feb 9;12:621564. doi: 10.3389/fmicb.2021.621564. eCollection 2021. Front Microbiol. 2021. PMID: 33633708 Free PMC article.
-
Acquisition of extended-spectrum beta-lactamase producing Escherichia coli strains in male and female infants admitted to a neonatal intensive care unit: molecular epidemiology and analysis of risk factors.J Med Microbiol. 2010 Aug;59(Pt 8):948-954. doi: 10.1099/jmm.0.020214-0. Epub 2010 Apr 29. J Med Microbiol. 2010. PMID: 20430903
-
Extended Spectrum Beta-Lactamase-Producing Gram-Negative Bacteria Recovered From an Amazonian Lake Near the City of Belém, Brazil.Front Microbiol. 2019 Feb 28;10:364. doi: 10.3389/fmicb.2019.00364. eCollection 2019. Front Microbiol. 2019. PMID: 30873145 Free PMC article.
Cited by
-
Mass bathing events in River Kshipra, Central India- influence on the water quality and the antibiotic susceptibility pattern of commensal E.coli.PLoS One. 2020 Mar 4;15(3):e0229664. doi: 10.1371/journal.pone.0229664. eCollection 2020. PLoS One. 2020. PMID: 32130236 Free PMC article.
-
A Three-Year Follow-Up Study of Antibiotic and Metal Residues, Antibiotic Resistance and Resistance Genes, Focusing on Kshipra-A River Associated with Holy Religious Mass-Bathing in India: Protocol Paper.Int J Environ Res Public Health. 2017 May 29;14(6):574. doi: 10.3390/ijerph14060574. Int J Environ Res Public Health. 2017. PMID: 28555050 Free PMC article.
-
Colistin Resistance Among Multiple Sequence Types of Klebsiella pneumoniae Is Associated With Diverse Resistance Mechanisms: A Report From India.Front Microbiol. 2021 Feb 22;12:609840. doi: 10.3389/fmicb.2021.609840. eCollection 2021. Front Microbiol. 2021. PMID: 33692764 Free PMC article.
-
Study of pandrug and heavy metal resistance among E. coli from anthropogenically influenced Delhi stretch of river Yamuna.Braz J Microbiol. 2018 Jul-Sep;49(3):471-480. doi: 10.1016/j.bjm.2017.11.001. Epub 2018 Feb 12. Braz J Microbiol. 2018. PMID: 29449175 Free PMC article.
-
Plasmid-Mediated Ampicillin, Quinolone, and Heavy Metal Co-Resistance among ESBL-Producing Isolates from the Yamuna River, New Delhi, India.Antibiotics (Basel). 2020 Nov 19;9(11):826. doi: 10.3390/antibiotics9110826. Antibiotics (Basel). 2020. PMID: 33227950 Free PMC article.
References
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

