Mifepristone Suppresses Basal Triple-Negative Breast Cancer Stem Cells by Down-regulating KLF5 Expression
- PMID: 26941846
- PMCID: PMC4775863
- DOI: 10.7150/thno.14315
Mifepristone Suppresses Basal Triple-Negative Breast Cancer Stem Cells by Down-regulating KLF5 Expression
Abstract
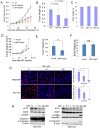
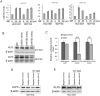
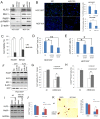
Triple-negative breast cancer (TNBC) is currently the most malignant subtype of breast cancers without effective targeted therapies. Mifepristone (MIF), a drug regularly used for abortion, has been reported to have anti-tumor activity in multiple hormone-dependent cancers, including luminal type breast cancers. In this study, we showed that MIF suppressed tumor growth of the TNBC cell lines and patient-derived xenografts in NOD-SCID mice. Furthermore, MIF reduced the TNBC cancer stem cell (CSC) population through down-regulating KLF5 expression, a stem cell transcription factor over-expressed in basal type TNBC and promoting cell proliferation, survival and stemness. Interestingly, MIF suppresses the expression of KLF5 through inducing the expression of miR-153. Consistently, miR-153 decreases CSC and miR-153 inhibitor rescued MIF-induced down-regulation of the KLF5 protein level and CSC ratio. Taken together, our findings suggest that MIF inhibits basal TNBC via the miR-153/KLF5 axis and MIF may be used for the treatment of TNBC.
Keywords: Cancer Stem Cell; KLF5; Mifepristone; Triple-negative Breast Cancer; miR-153..
Conflict of interest statement
Competing Interests: Authors have no conflicts of interest.
Figures



References
-
- Desmedt C, Ruiz-Garcia E, Andre F. Gene expression predictors in breast cancer: current status, limitations and perspectives. Eur J Cancer. 2008;44:2714–20. - PubMed
-
- Goldhirsch A, Wood WC, Coates AS. et al. Strategies for subtypes-dealing with the diversity of breast cancer: highlights of the St. Gallen International Expert Consensus on the Primary Therapy of Early Breast Cancer 2011. Annals of oncology: official journal of the European Society for Medical Oncology / ESMO. 2011;22:1736–47. - PMC - PubMed
-
- Dent R, Trudeau M, Pritchard KI. et al. Triple-negative breast cancer: clinical features and patterns of recurrence. Clinical cancer research: an official journal of the American Association for Cancer Research. 2007;13:4429–34. - PubMed
-
- Rakha EA, El-Sayed ME, Green AR. et al. Prognostic markers in triple-negative breast cancer. Cancer. 2007;109:25–32. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases
Miscellaneous

