PD-1 expression conditions T cell avidity within an antigen-specific repertoire
- PMID: 26942093
- PMCID: PMC4760290
- DOI: 10.1080/2162402X.2015.1104448
PD-1 expression conditions T cell avidity within an antigen-specific repertoire
Abstract
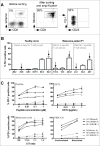
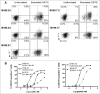
Despite its negative regulatory role on tumor-specific T cells, Programmed cell death 1 (PD-1) is also a marker of activated tumor-infiltrating T cells. In cancer, PD-1 blockade partially reverses T cell dysfunction allowing the amplification of tumor reactive T cells. Here, we investigated the role of PD-1 signaling on effector/memory human T cells specific for shared melanoma antigens, derived from blood. We documented for the first time the existence of melanoma-specific T cell clones unable to express PD-1. This stable feature was due to the persistent methylation of the PDCD1 promoter. These PD-1neg clones were of lower avidity than their PD-1pos counterparts, suggesting that high-affinity-specific T cell clones unable to express PD-1 are not or rarely present in peripheral blood, as they are probably eliminated by negative selection, due to their high reactivity. We also documented the existence of such PD-1neg T cell clones in melanoma tumor-infiltrating lymphocytes (TIL), which also exhibited a lower functional avidity than PD-1pos TIL clones. This clearly shows that PD-1 expression identifies antigen-specific T cell clonotypes of high functional avidity. Finally, we demonstrated that PD-1 blockade during the in vitro selection process of Melan-A-specific T cells favored the amplification of higher avidity T cell clonotypes. This preferential amplification of high-avidity memory T cells upon PD-1 blockade resonates with the expansion of reactive T cells, including neo-antigen-specific T cells observed in anti-PD-1-treated patients. This feature should also be a useful biomarker of clinical efficiency, while providing new insights for adoptive transfer treatments.
Keywords: Adoptive T cell transfer; Melan-A; PD-1; T cell avidity; immunotherapy; melanoma.
Figures




References
-
- Kamphorst AO, Ahmed R. Manipulating the PD-1 pathway to improve immunity. Curr Opin Immunol 2013; 25:381-8; PMID:23582509; http://dx.doi.org/10.1016/j.coi.2013.03.003 - DOI - PMC - PubMed
-
- Moll M, Kuylenstierna C, Gonzalez VD, Andersson SK, Bosnjak L, Sonnerborg A, Quigley MF, Sandberg JK. Severe functional impairment and elevated PD-1 expression in CD1d-restricted NKT cells retained during chronic HIV-1 infection. Eur J Immunol 2009; 39:902-11; PMID:19197939; http://dx.doi.org/10.1002/eji.200838780 - DOI - PMC - PubMed
-
- Riley JL. PD-1 signaling in primary T cells. Immunol Rev 2009; 229:114-25; PMID:19426218; http://dx.doi.org/10.1111/j.1600-065X.2009.00767.x - DOI - PMC - PubMed
-
- Nishimura H, Nose M, Hiai H, Minato N, Honjo T. Development of lupus-like autoimmune diseases by disruption of the PD-1 gene encoding an ITIM motif-carrying immunoreceptor. Immunity 1999; 11:141-51; PMID:10485649; http://dx.doi.org/10.1016/S1074-7613(00)80089-8 - DOI - PubMed
-
- Nishimura H, Okazaki T, Tanaka Y, Nakatani K, Hara M, Matsumori A, Sasayama S, Mizoguchi A, Hiai H, Minato N et al.. Autoimmune dilated cardiomyopathy in PD-1 receptor-deficient mice. Science 2001; 291:319-22; PMID:11209085; http://dx.doi.org/10.1126/science.291.5502.319 - DOI - PubMed
LinkOut - more resources
Full Text Sources
Other Literature Sources
