TRIM21 Ubiquitylates SQSTM1/p62 and Suppresses Protein Sequestration to Regulate Redox Homeostasis
- PMID: 26942676
- PMCID: PMC4779181
- DOI: 10.1016/j.molcel.2016.02.007
TRIM21 Ubiquitylates SQSTM1/p62 and Suppresses Protein Sequestration to Regulate Redox Homeostasis
Erratum in
-
TRIM21 Ubiquitylates SQSTM1/p62 and Suppresses Protein Sequestration to Regulate Redox Homeostasis.Mol Cell. 2016 Apr 7;62(1):149-51. doi: 10.1016/j.molcel.2016.03.015. Mol Cell. 2016. PMID: 27058791 No abstract available.
Abstract
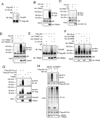
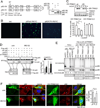
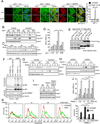
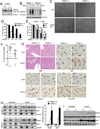
TRIM21 is a RING finger domain-containing ubiquitin E3 ligase whose expression is elevated in autoimmune disease. While TRIM21 plays an important role in immune activation during pathogen infection, little is known about its inherent cellular function. Here we show that TRIM21 plays an essential role in redox regulation by directly interacting with SQSTM1/p62 and ubiquitylating p62 at lysine 7 (K7) via K63-linkage. As p62 oligomerizes and sequesters client proteins in inclusions, the TRIM21-mediated p62 ubiquitylation abrogates p62 oligomerization and sequestration of proteins including Keap1, a negative regulator of antioxidant response. TRIM21-deficient cells display an enhanced antioxidant response and reduced cell death in response to oxidative stress. Genetic ablation of TRIM21 in mice confers protection from oxidative damages caused by arsenic-induced liver insult and pressure overload heart injury. Therefore, TRIM21 plays an essential role in p62-regulated redox homeostasis and may be a viable target for treating pathological conditions resulting from oxidative damage.
Copyright © 2016 Elsevier Inc. All rights reserved.
Figures


References
-
- Barchowsky A, Dudek EJ, Treadwell MD, Wetterhahn KE. Arsenic induces oxidant stress and NF-kappa B activation in cultured aortic endothelial cells. Free Radic Biol Med. 1996;21:783–790. - PubMed
-
- Ben-Chetrit E, Fox RI, Tan EM. Dissociation of immune responses to the SS-A (Ro) 52-kd and 60-kd polypeptides in systemic lupus erythematosus and Sjogren's syndrome. Arthritis and rheumatism. 1990;33:349–355. - PubMed
-
- Bernstam L, Nriagu J. Molecular aspects of arsenic stress. Journal of toxicology and environmental health Part B, Critical reviews. 2000;3:293–322. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- R01 DK108743/DK/NIDDK NIH HHS/United States
- R01 CA192642/CA/NCI NIH HHS/United States
- R01 GM097355/GM/NIGMS NIH HHS/United States
- R01 CA172025/CA/NCI NIH HHS/United States
- P30 CA030199/CA/NCI NIH HHS/United States
- R01CA192642/CA/NCI NIH HHS/United States
- R01 CA129536/CA/NCI NIH HHS/United States
- R01GM97355/GM/NIGMS NIH HHS/United States
- P30 CA072720/CA/NCI NIH HHS/United States
- 5P30CA030199/CA/NCI NIH HHS/United States
- R01DK108743/DK/NIDDK NIH HHS/United States
- R01CA172025/CA/NCI NIH HHS/United States
- R01CA129536/CA/NCI NIH HHS/United States
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials

