Cancer therapeutic approach based on conformational stabilization of mutant p53 protein by small peptides
- PMID: 26943582
- PMCID: PMC4914250
- DOI: 10.18632/oncotarget.7857
Cancer therapeutic approach based on conformational stabilization of mutant p53 protein by small peptides
Abstract
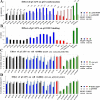
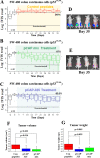
The p53 tumor suppressor serves as a major barrier against malignant transformation. Over 50% of tumors inactivate p53 by point mutations in its DNA binding domain. Most mutations destabilize p53 protein folding, causing its partial denaturation at physiological temperature. Thus a high proportion of human tumors overexpress a potential potent tumor suppressor in a non-functional, misfolded form. The equilibrium between the properly folded and misfolded states of p53 may be affected by molecules that interact with p53, stabilizing its native folding and restoring wild type p53 activity to cancer cells. To select for mutant p53 (mutp53) reactivating peptides, we adopted the phage display technology, allowing interactions between mutp53 and random peptide libraries presented on phages and enriching for phage that favor the correctly folded p53 conformation. We obtained a large database of potential reactivating peptides. Lead peptides were synthesized and analyzed for their ability to restore proper p53 folding and activity. Remarkably, many enriched peptides corresponded to known p53-binding proteins, including RAD9. Importantly, lead peptides elicited dramatic regression of aggressive tumors in mouse xenograft models. Such peptides might serve as novel agents for human cancer therapy.
Keywords: conformation; p53; peptides; pre-clinical; reactivation.
Conflict of interest statement
The authors are supported by a Center of Excellence of the Flight Attendant Medical Research Institute (FAMRI) and by an Israel Science Foundation (ISF) Center of Excellence. The terms of this arrangement have been reviewed and approved by the Weizmann Institute of Science in accordance with its policy on objectivity in research.
Figures




References
-
- Levine JA. p53, the cellular gatekeeper for growth and division. Cell. 1997;88:323–331. - PubMed
-
- Vousden KH, Lu X. Live or let die: the cell's response to p53. Nat Rev Cancer. 2002;2:594–604. - PubMed
-
- el-Deiry WS, Kern SE, Pietenpol JA, Kinzler KW, Vogelstein B. Definition of a consensus binding site for p53. Nat Genet. 1992;1:45–49. - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
Miscellaneous

