Metagenomic Signatures of Bacterial Adaptation to Life in the Phyllosphere of a Salt-Secreting Desert Tree
- PMID: 26944845
- PMCID: PMC4836421
- DOI: 10.1128/AEM.00483-16
Metagenomic Signatures of Bacterial Adaptation to Life in the Phyllosphere of a Salt-Secreting Desert Tree
Abstract
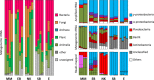
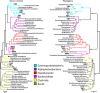
The leaves of Tamarix aphylla, a globally distributed, salt-secreting desert tree, are dotted with alkaline droplets of high salinity. To successfully inhabit these organic carbon-rich droplets, bacteria need to be adapted to multiple stress factors, including high salinity, high alkalinity, high UV radiation, and periodic desiccation. To identify genes that are important for survival in this harsh habitat, microbial community DNA was extracted from the leaf surfaces of 10 Tamarix aphylla trees along a 350-km longitudinal gradient. Shotgun metagenomic sequencing, contig assembly, and binning yielded 17 genome bins, six of which were >80% complete. These genomic bins, representing three phyla (Proteobacteria,Bacteroidetes, and Firmicutes), were closely related to halophilic and alkaliphilic taxa isolated from aquatic and soil environments. Comparison of these genomic bins to the genomes of their closest relatives revealed functional traits characteristic of bacterial populations inhabiting the Tamarix phyllosphere, independent of their taxonomic affiliation. These functions, most notably light-sensing genes, are postulated to represent important adaptations toward colonization of this habitat.
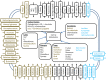
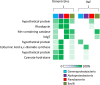
Importance: Plant leaves are an extensive and diverse microbial habitat, forming the main interface between solar energy and the terrestrial biosphere. There are hundreds of thousands of plant species in the world, exhibiting a wide range of morphologies, leaf surface chemistries, and ecological ranges. In order to understand the core adaptations of microorganisms to this habitat, it is important to diversify the type of leaves that are studied. This study provides an analysis of the genomic content of the most abundant bacterial inhabitants of the globally distributed, salt-secreting desert tree Tamarix aphylla Draft genomes of these bacteria were assembled, using the culture-independent technique of assembly and binning of metagenomic data. Analysis of the genomes reveals traits that are important for survival in this habitat, most notably, light-sensing and light utilization genes.
Copyright © 2016, American Society for Microbiology. All Rights Reserved.
Figures


References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

