Gene expression profiling of the venom gland from the Venezuelan mapanare (Bothrops colombiensis) using expressed sequence tags (ESTs)
- PMID: 26944950
- PMCID: PMC4779267
- DOI: 10.1186/s12867-016-0059-7
Gene expression profiling of the venom gland from the Venezuelan mapanare (Bothrops colombiensis) using expressed sequence tags (ESTs)
Erratum in
-
Erratum to: Gene expression profiling of the venom gland from the Venezuelan mapanare (Bothrops colombiensis) using expressed sequence tags (ESTs).BMC Mol Biol. 2016 May 24;17(1):13. doi: 10.1186/s12867-016-0062-z. BMC Mol Biol. 2016. PMID: 27220910 Free PMC article. No abstract available.
Abstract
Background: Bothrops colombiensis is a highly dangerous pit viper and responsible for over 70% of snakebites in Venezuela. Although the composition in B. colombiensis venom has been identified using a proteome analysis, the venom gland transcriptome is currently lacking.
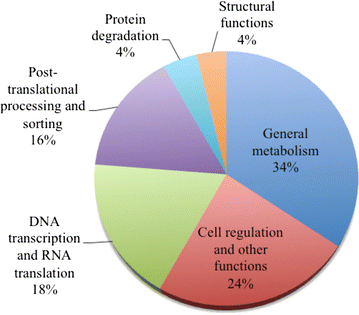
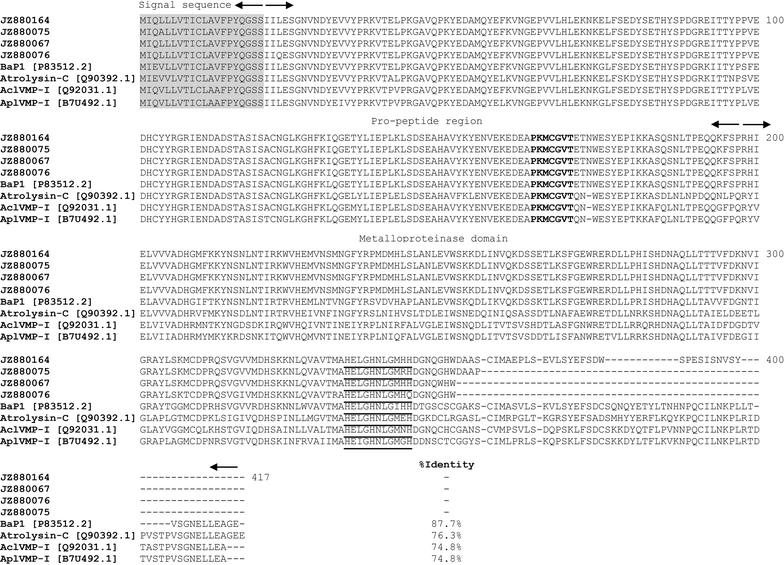
Results: We constructed a cDNA library from the venom gland of B. colombiensis, and a set of 729 high quality expressed sequence tags (ESTs) was identified. A total number of 344 ESTs (47.2% of total ESTs) was related to toxins. The most abundant toxin transcripts were metalloproteinases (37.5%), phospholipases A2s (PLA2, 29.7%), and serine proteinases (11.9%). Minor toxin transcripts were linked to waprins (5.5%), C-type lectins (4.1%), ATPases (2.9%), cysteine-rich secretory proteins (CRISP, 2.3%), snake venom vascular endothelium growth factors (svVEGF, 2.3%), L-amino acid oxidases (2%), and other putative toxins (1.7%). While 160 ESTs (22% of total ESTs) coded for translation proteins, regulatory proteins, ribosomal proteins, elongation factors, release factors, metabolic proteins, and immune response proteins. Other proteins detected in the transcriptome (87 ESTs, 11.9% of total ESTs) were undescribed proteins with unknown functions. The remaining 138 (18.9%) cDNAs had no match with known GenBank accessions.
Conclusion: This study represents the analysis of transcript expressions and provides a physical resource of unique genes for further study of gene function and the development of novel molecules for medical applications.
Figures






References
-
- Giron ME, Rodriguez-Acosta A, Salazar AM, Sanchez EE, Galan J, Ibarra C, et al. Isolation and characterization of two new non-hemorrhagic metalloproteinases with fibrinogenolytic activity from the mapanare (Bothrops colombiensis) venom. Arch Toxicol. 2013;87(1):197–208. doi: 10.1007/s00204-012-0914-3. - DOI - PubMed
-
- Giron ME, Salazar AM, Aguilar I, Perez JC, Sanchez EE, Arocha-Pinango CL, et al. Hemorrhagic, coagulant and fibrino(geno)lytic activities of crude venom and fractions from mapanare (Bothrops colombiensis) snakes. Comp Biochem Physiol C: Toxicol Pharmacol. 2008;147(1):113–121. - PubMed
-
- de Selistre Araujo HS, de Souza DH, Ownby CL. Analysis of a cDNA sequence encoding a novel member of the snake venom metalloproteinase, disintegrin-like, cysteine-rich (MDC) protein family from Agkistrodon contortrix laticinctus. Biochim Biophys Acta. 1997;1342(2):109–115. doi: 10.1016/S0167-4838(97)00111-8. - DOI - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

