Structure and function of the AAA+ ATPase p97/Cdc48p
- PMID: 26945625
- PMCID: PMC4821690
- DOI: 10.1016/j.gene.2016.02.042
Structure and function of the AAA+ ATPase p97/Cdc48p
Abstract
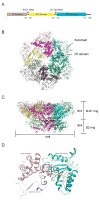
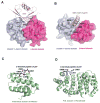
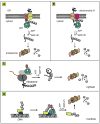
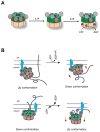
p97 (also known as valosin-containing protein (VCP) in mammals or Cdc48p in Saccharomyces cerevisiae) is an evolutionarily conserved ATPase present in all eukaryotes and archaebacteria. In conjunction with a collection of cofactors and adaptors, p97/Cdc48p performs an array of biological functions mostly through modulating the stability of 'client' proteins. Using energy from ATP hydrolysis, p97/Cdc48p segregates these molecules from immobile cellular structures such as protein assemblies, membrane organelles, and chromatin. Consequently, the released polypeptides can be efficiently degraded by the ubiquitin proteasome system or recycled. This review summarizes our current understanding of the structure and function of this essential cellular chaperoning system.
Keywords: AAA+ ATPase; Chromatin-associated degradation; ER-associated degradation; Membrane fusion; Mitochondria-associated degradation; Proteasome; Protein quality control; Ribosome-associated degradation; Segregase; Ubiquitin; Unfoldase; p97/CDC48.
Published by Elsevier B.V.
Figures

References
-
- Acharya U, Jacobs R, Peters JM, Watson N, Farquhar MG, Malhotra V. The formation of Golgi stacks from vesiculated Golgi membranes requires two distinct fusion events. Cell. 1995;82:895–904. - PubMed
-
- Acs K, Luijsterburg MS, Ackermann L, Salomons FA, Hoppe T, Dantuma NP. The AAA-ATPase VCP/p97 promotes 53BP1 recruitment by removing L3MBTL1 from DNA double-strand breaks. Nature structural & molecular biology. 2011;18:1345–1350. - PubMed
-
- Allen MD, Buchberger A, Bycroft M. The PUB domain functions as a p97 binding module in human peptide N-glycanase. The Journal of biological chemistry. 2006;281:25502–25508. - PubMed
-
- Anderson DJ, Le Moigne R, Djakovic S, Kumar B, Rice J, Wong S, Wang J, Yao B, Valle E, Kiss von Soly S, Madriaga A, Soriano F, Menon MK, Wu ZY, Kampmann M, Chen Y, Weissman JS, Aftab BT, Yakes FM, Shawver L, Zhou HJ, Wustrow D, Rolfe M. Targeting the AAA ATPase p97 as an Approach to Treat Cancer through Disruption of Protein Homeostasis. Cancer cell. 2015;28:653–665. - PMC - PubMed
-
- Ballar P, Shen Y, Yang H, Fang S. The role of a novel p97/valosin-containing protein-interacting motif of gp78 in endoplasmic reticulum-associated degradation. The Journal of biological chemistry. 2006;281:35359–35368. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous

