Data publication with the structural biology data grid supports live analysis
- PMID: 26947396
- PMCID: PMC4786681
- DOI: 10.1038/ncomms10882
Data publication with the structural biology data grid supports live analysis
Abstract
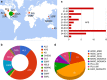
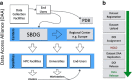
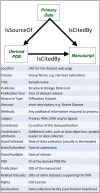
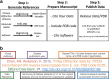
Access to experimental X-ray diffraction image data is fundamental for validation and reproduction of macromolecular models and indispensable for development of structural biology processing methods. Here, we established a diffraction data publication and dissemination system, Structural Biology Data Grid (SBDG; data.sbgrid.org), to preserve primary experimental data sets that support scientific publications. Data sets are accessible to researchers through a community driven data grid, which facilitates global data access. Our analysis of a pilot collection of crystallographic data sets demonstrates that the information archived by SBDG is sufficient to reprocess data to statistics that meet or exceed the quality of the original published structures. SBDG has extended its services to the entire community and is used to develop support for other types of biomedical data sets. It is anticipated that access to the experimental data sets will enhance the paradigm shift in the community towards a much more dynamic body of continuously improving data analysis.
Figures



References
-
- Bilderback D. H., Elleaume P. & Weckert E. Review of third and next generation synchrotron light sources. J. Phys. B: At. Mol. Opt. Phys. 38, S773–S797 (2005).
Publication types
MeSH terms
Substances
Grants and funding
- T32 GM008320/GM/NIGMS NIH HHS/United States
- U19 AI107792/AI/NIAID NIH HHS/United States
- U54 EB020406/EB/NIBIB NIH HHS/United States
- R01 GM065484/GM/NIGMS NIH HHS/United States
- R01 GM086487/GM/NIGMS NIH HHS/United States
- HHMI/Howard Hughes Medical Institute/United States
- R01 HL119813/HL/NHLBI NIH HHS/United States
- R01 GM117299/GM/NIGMS NIH HHS/United States
- R01 GM104141/GM/NIGMS NIH HHS/United States
- R01 GM117080/GM/NIGMS NIH HHS/United States
- U54 HL112303/HL/NHLBI NIH HHS/United States
- R01 CA140522/CA/NCI NIH HHS/United States
- NIH 1U54EB020406-01/EB/NIBIB NIH HHS/United States
- R01 NS085078/NS/NINDS NIH HHS/United States
- R21 GM110580/GM/NIGMS NIH HHS/United States
- 101908/Z/13/Z/WT_/Wellcome Trust/United Kingdom
- S10 RR028832/RR/NCRR NIH HHS/United States
- P41 GM103403/GM/NIGMS NIH HHS/United States
- R01 GM088454/GM/NIGMS NIH HHS/United States
- R21 AI065886/AI/NIAID NIH HHS/United States
- R01 GM069857/GM/NIGMS NIH HHS/United States
- R01 GM053163/GM/NIGMS NIH HHS/United States
- P41 GM103485/GM/NIGMS NIH HHS/United States
- R01 CA163647/CA/NCI NIH HHS/United States
- R01 MH077303/MH/NIMH NIH HHS/United States
- R01 GM112973/GM/NIGMS NIH HHS/United States
- R01 GM114358/GM/NIGMS NIH HHS/United States
- R01 GM110352/GM/NIGMS NIH HHS/United States
- R01 GM087353/GM/NIGMS NIH HHS/United States
- R01 AI045937/AI/NIAID NIH HHS/United States
- DP2 OD001996/OD/NIH HHS/United States
- NIH P41 GM103403/GM/NIGMS NIH HHS/United States
- 1S10RR028832/RR/NCRR NIH HHS/United States
LinkOut - more resources
Full Text Sources
Other Literature Sources

