The Complete Genome Sequences, Unique Mutational Spectra, and Developmental Potency of Adult Neurons Revealed by Cloning
- PMID: 26948891
- PMCID: PMC4795965
- DOI: 10.1016/j.neuron.2016.02.004
The Complete Genome Sequences, Unique Mutational Spectra, and Developmental Potency of Adult Neurons Revealed by Cloning
Abstract
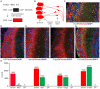
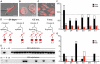
Somatic mutation in neurons is linked to neurologic disease and implicated in cell-type diversification. However, the origin, extent, and patterns of genomic mutation in neurons remain unknown. We established a nuclear transfer method to clonally amplify the genomes of neurons from adult mice for whole-genome sequencing. Comprehensive mutation detection and independent validation revealed that individual neurons harbor ∼100 unique mutations from all classes but lack recurrent rearrangements. Most neurons contain at least one gene-disrupting mutation and rare (0-2) mobile element insertions. The frequency and gene bias of neuronal mutations differ from other lineages, potentially due to novel mechanisms governing postmitotic mutation. Fertile mice were cloned from several neurons, establishing the compatibility of mutated adult neuronal genomes with reprogramming to pluripotency and development.
Copyright © 2016 Elsevier Inc. All rights reserved.
Figures


Comment in
-
Tracking Down Mutations Cell by Cell.Neuron. 2016 Mar 16;89(6):1126-1127. doi: 10.1016/j.neuron.2016.02.042. Neuron. 2016. PMID: 26985720
References
-
- Beale RCL, Petersen-Mahrt SK, Watt IN, Harris RS, Rada C, Neuberger MS. Comparison of the Differential Context-dependence of DNA Deamination by APOBEC Enzymes: Correlation with Mutation Spectra in Vivo. Journal of Molecular Biology. 2004;337:585–596. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
