Maturation Pathway from Germline to Broad HIV-1 Neutralizer of a CD4-Mimic Antibody
- PMID: 26949186
- PMCID: PMC4826291
- DOI: 10.1016/j.cell.2016.02.022
Maturation Pathway from Germline to Broad HIV-1 Neutralizer of a CD4-Mimic Antibody
Abstract
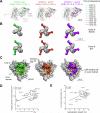
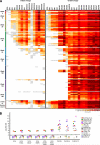
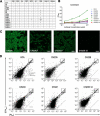
Antibodies with ontogenies from VH1-2 or VH1-46-germline genes dominate the broadly neutralizing response against the CD4-binding site (CD4bs) on HIV-1. Here, we define with longitudinal sampling from time-of-infection the development of a VH1-46-derived antibody lineage that matured to neutralize 90% of HIV-1 isolates. Structures of lineage antibodies CH235 (week 41 from time-of-infection, 18% breadth), CH235.9 (week 152, 77%), and CH235.12 (week 323, 90%) demonstrated the maturing epitope to focus on the conformationally invariant portion of the CD4bs. Similarities between CH235 lineage and five unrelated CD4bs lineages in epitope focusing, length-of-time to develop breadth, and extraordinary level of somatic hypermutation suggested commonalities in maturation among all CD4bs antibodies. Fortunately, the required CH235-lineage hypermutation appeared substantially guided by the intrinsic mutability of the VH1-46 gene, which closely resembled VH1-2. We integrated our CH235-lineage findings with a second broadly neutralizing lineage and HIV-1 co-evolution to suggest a vaccination strategy for inducing both lineages.
Copyright © 2016 Elsevier Inc. All rights reserved.
Figures



References
-
- Alam SM, McAdams M, Boren D, Rak M, Scearce RM, Gao F, Camacho ZT, Gewirth D, Kelsoe G, Chen P, et al. The role of antibody polyspecificity and lipid reactivity in binding of broadly neutralizing anti-HIV-1 envelope human monoclonal antibodies 2F5 and 4E10 to glycoprotein 41 membrane proximal envelope epitopes. J. Immunol. 2007;178:4424–4435. - PMC - PubMed
-
- Bonsignori M, Hwang KK, Chen X, Tsao CY, Morris L, Gray E, Marshall DJ, Crump JA, Kapiga SH, Sam NE, et al. Analysis of a clonal lineage of HIV-1 envelope V2/V3 conformational epitope-specific broadly neutralizing antibodies and their inferred unmutated common ancestors. J. Virol. 2011;85:9998–10009. - PMC - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Research Materials
Miscellaneous

