No current evidence for widespread dosage compensation in S. cerevisiae
- PMID: 26949255
- PMCID: PMC4798953
- DOI: 10.7554/eLife.10996
No current evidence for widespread dosage compensation in S. cerevisiae
Abstract
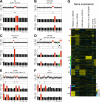
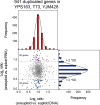
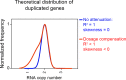
Previous studies of laboratory strains of budding yeast had shown that when gene copy number is altered experimentally, RNA levels generally scale accordingly. This is true when the copy number of individual genes or entire chromosomes is altered. In a recent study, Hose et al. (2015) reported that this tight correlation between gene copy number and RNA levels is not observed in recently isolated wild Saccharomyces cerevisiae variants. To understand the origins of this proposed difference in gene expression regulation between natural variants and laboratory strains of S. cerevisiae, we evaluated the karyotype and gene expression studies performed by Hose et al. on wild S. cerevisiae strains. In contrast to the results of Hose et al., our reexamination of their data revealed a tight correlation between gene copy number and gene expression. We conclude that widespread dosage compensation occurs neither in laboratory strains nor in natural variants of S. cerevisiae.
Keywords: S. cerevisiae; aneuploidy; cell biology; chromosomes; dosage compensation; gene expression; genes.
Conflict of interest statement
The authors declare that no competing interests exist.
Figures





References
-
- Dabeva MD, Warner JR. The yeast ribosomal protein L32 and its gene. The Journal of Biological Chemistry. 1987;262:16055–16059. - PubMed
-
- Dunham MJ, Badrane H, Ferea T, Adams J, Brown PO, Rosenzweig F, Botstein D. Characteristic genome rearrangements in experimental evolution of saccharomyces cerevisiae. Proceedings of the National Academy of Sciences of the United States of America. 2002;99:16144–16149. doi: 10.1073/pnas.242624799. - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

