Searching for Drug Synergy in Complex Dose-Response Landscapes Using an Interaction Potency Model
- PMID: 26949479
- PMCID: PMC4759128
- DOI: 10.1016/j.csbj.2015.09.001
Searching for Drug Synergy in Complex Dose-Response Landscapes Using an Interaction Potency Model
Erratum in
-
Corrigendum to "Searching for drug synergy in complex dose-response landscapes using an interaction potency model" [Comput. Struct. Biotechnol. J. 13 (2015) 504-513].Comput Struct Biotechnol J. 2017 Jul 25;15:387. doi: 10.1016/j.csbj.2017.07.003. eCollection 2017. Comput Struct Biotechnol J. 2017. PMID: 29991977 Free PMC article.
Abstract
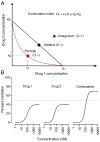
Rational design of multi-targeted drug combinations is a promising strategy to tackle the drug resistance problem for many complex disorders. A drug combination is usually classified as synergistic or antagonistic, depending on the deviation of the observed combination response from the expected effect calculated based on a reference model of non-interaction. The existing reference models were proposed originally for low-throughput drug combination experiments, which make the model assumptions often incompatible with the complex drug interaction patterns across various dose pairs that are typically observed in large-scale dose-response matrix experiments. To address these limitations, we proposed a novel reference model, named zero interaction potency (ZIP), which captures the drug interaction relationships by comparing the change in the potency of the dose-response curves between individual drugs and their combinations. We utilized a delta score to quantify the deviation from the expectation of zero interaction, and proved that a delta score value of zero implies both probabilistic independence and dose additivity. Using data from a large-scale anticancer drug combination experiment, we demonstrated empirically how the ZIP scoring approach captures the experimentally confirmed drug synergy while keeping the false positive rate at a low level. Further, rather than relying on a single parameter to assess drug interaction, we proposed the use of an interaction landscape over the full dose-response matrix to identify and quantify synergistic and antagonistic dose regions. The interaction landscape offers an increased power to differentiate between various classes of drug combinations, and may therefore provide an improved means for understanding their mechanisms of action toward clinical translation.
Keywords: Dose–response matrix; Drug combination scoring; High-throughput screening; Interaction landscape.
Figures





References
LinkOut - more resources
Full Text Sources
Other Literature Sources
